Abstract
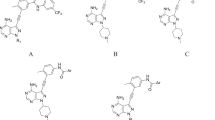
Anaplastic lymphoma kinase (ALK) is involved in many signaling mechanisms that lead to cell-cycle progression; overexpression of ALK has been found in many types of cancers. ALK is a recognized target for the development of small-molecule inhibitors for the treatment of cancer. In this study, a diverse set of 71 ALK inhibitors were aligned by three different methods (pharmacophore, docking-based, and rigid body (Distill) alignment) for the development of comparative molecular field analysis (CoMFA) and comparative molecular similarity indices analysis (CoMSIA) models. The best 3D QSAR models were obtained, which used rigid body (Distill) alignment of test and training set molecules. CoMFA and CoMSIA models were found statistically significant with leave-one-out correlation coefficients (q 2) of 0.816 and 0.838, respectively; cross-validated coefficients (\(r_{\text{cv}}^{2}\)) of 0.812 and 0.837, respectively; and conventional coefficients (r 2) of 0.969 and 0.966, respectively. QSAR models were validated by a test set of 14 compounds giving satisfactory prediction of correlation coefficients (\(r_{\text{pred}}^{2}\)) of 0.910 and 0.904 for CoMFA and CoMSIA models, respectively. Based on the generated contour maps, we have designed 10 novel ALK inhibitors and predicted their activities. Finally, molecular docking study was performed for designed molecules. The designed compounds showed good potential to be used as ALK inhibitors.






Similar content being viewed by others
References
Allwein SP, Roemmele RC, Haley JJ, Mowrey DR, Petrillo DE, Reif JJ, Gingrich DE, Bakale RP (2012) Development and scale-up of an optimized route to the ALK inhibitor CEP-28122. Org Process Res Dev 16:148–155
Bryan MC, Whittington DA, Doherty EM, Falsey JR, Cheng AC, Emkey R, Brake RL, Lewis RT (2012) Rapid development of piperidine carboxamides as potent and selective anaplastic lymphoma kinase inhibitors. J Med Chem 55:1698–1705
Cheng M, Ott GR (2010) Anaplastic lymphoma kinase as a therapeutic target in anaplastic large cell lymphoma, non-small cell lung cancer and neuroblastoma. Anti-Cancer Agents Med Chem 10:236–249
Clark M, Cramer RD, Opdenbosch NV (1989) Validation of the general purpose tripos 5.2 force field. J Comput Chem 10:982–1012
Cramer RD III, Bunce JD, Patterson DE (1988) Crossvalidation, bootstrapping, and partial least squares compared with multiple regression in conventional QSAR studies. Quant Struct Act Relat 7:18–25
Gasteiger J, Marsili M (1980) Iterative partial equalization of orbital electronegativity—a rapid access to atomic charges. Tetrahedron 36:3219–3228
Jain AN (1996) Scoring non-covalent protein-ligand interactions: a continuous differentiable function tuned to compute binding affinities. J Comput Aided-Mol Des 10:427–440
Klebe G, Abraham U (1999) Comparative molecular similarity index analysis (CoMSIA) to study hydrogen-bonding properties and to score combinatorial libraries. J Comput Aided Mol Des 13:1–10
Lee C (2010) Crystal structure of the ALK (anaplastic lymphoma kinase) catalytic domain. Biochem J 430:425–437
Lewis RT, Bode CM, Choquette DM, Potashman M, Romero K, Stellwagen JC, Teffera Y, Moore E, Whittington DA, Chen H, Epstein LF, Emkey R, Andrews PS, Yu VL, Saffran DC, Xu M, Drew A, Merkel P, Szilvassy S, Brake RL (2012) The discovery and optimization of a novel class of potent, selective, and orally bioavailable anaplastic lymphoma kinase (ALK) inhibitors with potential utility for the treatment of cancer. J Med Chem 55:6523–6540
Moh K, Saito N, Okabe T, Masuda Y, Furuya T, Nagano T (2011) Virtual screening and further development of novel ALK inhibitors. Bioorg Med Chem 19:3086–3095
Palmer RH, Vernersson E, Grabbe C, Hallberg B (2009) Anaplastic lymphoma kinase: signalling in development and disease. Biochem J 15:345–361
Pulford K, Morris SW, Turturro F (2004) Anaplastic lymphoma kinase proteins in growth control and cancer. J Cell Physiol 199:330–358
Roy KK, Dixit A, Saxena AK (2008) An investigation of structurally diverse carbamates for acetylcholinesterase (AChE) inhibition using 3D-QSAR analysis. J Mol Graphics Model 27:197–208
Shaw AT, Yasothan U, Kirkpatrick P (2011) Crizotinib. Nature Rev Drug Discov 10:897–898
Tartari CJ, Scapozza L, Gambacorti-Passerini C (2011) The ALK gene an attractive target for inhibitor development. Curr Top Med Chem 11:1406–1419
Tripathy R, McHugh RJ, Ghose AK, Ott GR, Angeles TS, Albom MS, Huang Z, Aimone LD, Cheng M, Dorsey BD (2011) Pyrazolone-based anaplastic lymphoma kinase (ALK) inhibitors: control of selectivity by a benzyloxy group. Bioorg Med Chem Lett 21:7261–7264
Vyas VK, Ghate M (2012a) 2D and 3D QSAR study on amino nicotinic acid and isonicotinic acid derivatives as potential inhibitors of dihydroorotate dehydrogenase (DHODH). Med Chem Res 21:3021–3034
Vyas VK, Ghate M (2012b) CoMFA and CoMSIA studies on aryl carboxylic acid amide derivatives as dihydroorotate dehydrogenase (DHODH) inhibitors. Cur Comp Aided-Drug Des 8:271–282
Vyas VK, Bhatt HG, Patel PK, Jalu J, Chintha C, Gupta N, Ghate M (2012) CoMFA and CoMSIA studies on C-aryl glucoside SGLT2 inhibitors as potential antidiabetic agents. SAR QSAR Environ Res. doi:10.1080/1062936X.2012.751553
Vyas VK, Ghate M, Parikh H (2013) 3D QSAR studies on 5-(2-methylbenzimidazol-1-yl)-N-alkylthiophene-2-carboxamide derivatives as P. falciparum dihydroorotate dehydrogenase (PfDHODH) inhibitors. Med Chem Res. doi:10.1007/s00044-012-0216-6
Zificsak CA, Theroff JP, Aimone LD, Angeles TS, Albom MS, Cheng M, Mesaros EF, Ott GR, Quail MR, Underiner TL, Wan W, Dorsey BD (2011) Methanesulfonamido-cyclohexylamine derivatives of 2,4-diaminopyrimidine as potent ALK inhibitors. Bioorg Med Chem Lett 21:3877–3880
Acknowledgments
The authors would like to thank Nirma University, Ahmedabad, India for providing the necessary facilities.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Vyas, V.K., Patel, A., Gupta, N. et al. Design of novel anaplastic lymphoma kinase (ALK) inhibitors based on predictive 3D QSAR models using different alignment strategies. Med Chem Res 23, 603–617 (2014). https://doi.org/10.1007/s00044-013-0662-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00044-013-0662-9