Abstract
Cell fusion of female and male gametes is the climax of sexual reproduction. In many organisms, the Hapless 2 (HAP2) family of proteins play a critical role in gamete fusion. We find that Plasmodium falciparum, the causative agent of human malaria, expresses two HAP2 proteins: PfHAP2 and PfHAP2p. These proteins are present in stage V gametocytes and localize throughout the flagellum of male gametes. Gene deletion analysis and genetic crosses show that PfHAP2 and PfHAP2p individually are essential for male fertility and thereby, parasite transmission to the mosquito. Using a cell fusion assay, we demonstrate that PfHAP2 and PfHAP2p are both authentic plasma membrane fusogens. Our results establish nonredundant essential roles for PfHAP2 and PfHAP2p in mediating gamete fusion in Plasmodium and suggest avenues in the design of novel strategies to prevent malaria parasite transmission from humans to mosquitoes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Malaria remains a major global health burden and a major cause of mortality and morbidity in developing countries worldwide. It is caused by Plasmodium parasites, with most deaths attributed to infection with Plasmodium falciparum. Malaria parasites are haploid Alveolates that reproduce asexually within two hosts: a vertebrate such as humans and a mosquito. Transmission from a human host to a mosquito vector requires sexual reproduction, which is essential for continuing the parasite life cycle. Sexual reproduction creates genetic diversity via recombination, allowing the parasite to rapidly adapt to its host and vector environment. Yet, sexual reproduction also constitutes a population bottleneck in the parasite life cycle that is vulnerable to disruption and the target of vaccine development efforts [1]. Plasmodium sexual stages form from a subset of asexually replicating parasites in the human blood stream and in P. falciparum, develop over a period of approximately 2 weeks into mature male and female gametocytes. They are then taken up by Anopheles mosquitoes during blood meal acquisition. Inside the mosquito midgut lumen, gametocytes are activated, rapidly differentiate into gametes, and egress from the infected red blood cells [2,3,4]. The male gametocyte undergoes three rapid rounds of DNA replication and forms eight flagellar (male) microgametes during its maturation [5]. The female gametocyte undergoes a marked reduction in cytoplasmic density and nuclear changes to form a single macrogamete [2, 3]. Male gametes exhibit motility through the blood meal and when encountering female gametes, attach and fuse, forming a short-lived diploid zygote [3, 5]. Zygotes differentiate into motile tetraploid ookinetes [6], penetrate the mosquito midgut epithelium and develop into oocysts. Within 2–3 weeks, oocysts differentiate into haploid invasive sporozoites [7], which migrate to the salivary glands for transmission to a new host.
Gamete interactions that precede cell fusion involve four members of the Apicomplexan specific s48/45 domain 6-cysteine (6-cys) family of proteins—P48/45, P47, P230 and P230p [8,9,10,11,12]. Fertilization of the macrogamete by the microgamete then requires cell fusion. Yet, the molecular mechanisms underlying gamete fusion in Plasmodium and the involved fusogens are poorly understood. Eukaryotes utilize a wide array of proteins for different stages in fertilization, which can be distinct between vertebrates, invertebrates, plants, and protozoans [13,14,15,16]. One family of proteins involved in fertilization are the Hapless 2 (HAP2)/Generative Cell Specific 1 (GCS1) proteins which were identified in genetic screens [17, 18]. Moreover, HAP2(GCS1) from Arabidopsis was shown to be an authentic cell fusogen that can fuse heterologous cells [19,20,21]. HAP2 proteins show structural similarity to Class II viral fusion proteins and somatic animal fusogens [19–21]. These viral, somatic, and sexual fusogens are members of the fusexin superfamily and have a large extracellular region containing three conserved globular domains (domain I, domain II and domain III) [19]. As in viral fusogens, a conserved, ~ 46 amino acid residue region in domain II of HAP2 (the cd-loop) has been suggested to be involved in the fusion process by inserting itself into target membranes [19,20,21]. HAP2 gene knockouts in Chlamydomonas resulted in gametes of opposing mating types—mt(+) and mt(−)—to adhere to each other, but membrane fusion and thus fertilization was abrogated [22]. In the apicomplexan parasite Toxoplasma gondii, HAP2 also has a role in fertilization, and TgHAP2 knockout gametes show reduced fertilization [23]. Furthermore, two studies using the rodent malaria parasite Plasmodium berghei identified PbHAP2 and showed that it has a critical role in fertilization [22, 24]. However, the cell membrane fusogenic activity of PbHAP2 was not demonstrated.
In search for putative gamete fusogens of Plasmodium, we found that the human malaria parasite P. falciparum, as well as all other examined Plasmodium species, harbor two HAP2-like protein-encoding genes; the canonical PfHAP2 and a paralog, PfHAP2p. We show that in P. falciparum, HAP2 and HAP2p are expressed in gametocytes, activated male gametocytes, and microgametes. Interestingly, PfHAP2p was also expressed by females. In the absence of either protein, fertilization and transmission of the parasite to mosquitoes was completely blocked. Importantly, independent expression of PfHAP2 or PfHAP2p in mammalian cells mediated cell fusion, providing the first direct experimental evidence that these proteins serve as authentic cell membrane fusogens during Plasmodium sexual reproduction. This suggests PfHAP2 and PfHAP2p as attractive targets for developing transmission-blocking strategies against the most important human malaria parasites.
Results
Two HAP2/GCS1-like proteins are encoded in Plasmodium genomes.
To identify genes encoding HAP2/GCS1-like proteins in Plasmodium genomes, we searched for the HAP2 (PFAM 10699) domain in the P. falciparum genome using PlasmoDB version 50 (https://plasmodb.org/plasmo/app). This revealed two distinct HAP2/GCS1-encoding genes in all Plasmodium species, PfHAP2 (PlasmoDB ID PF3D7_1014200) and PfHAP2p (paralog of HAP2, PlasmoDB ID PF3D7_0816300). In P. falciparum these are located on different chromosomes (chr.), namely PfHAP2 on chr. 10 and PfHAP2p on chr. 8. Domain analysis revealed a conserved domain architecture for both PfHAP2 and PfHAP2p (Fig. 1A, B), however, PfHAP2p lacked a predicted N-terminal signal peptide and possessed a longer cytoplasmic domain (Fig. 1A, B). HAP2 proteins showed structural similarity to eukaryotic/viral class II fusion proteins, particularly their domain II (DII) was predicted to contain a ~ 42-residue cd-fusion loop [19]. An amino acid sequence alignment of a part of DII regions from different HAP2 proteins: P. falciparum (PfHAP2 and PfHAP2p), Chlamydomonas reinhardtii (Cr), Leishmania major (Lm), Tribolum castaneum (Tc), Tetrahymena thermophile (Tt), Arabidopsis thaliana (At), Toxoplasma gondii (Tg), and Cryptosporidium muris (Cm), revealed conservation of several residues in the predicted cd-fusion loops in PfHAP2 and PfHAP2p which may be relevant to disulfide bridge formation, but multiple sequence differences were observed in different species (Fig. 1C). Sequence alignment of the HAP2 domain (PFAM10699) of PfHAP2 and PfHAP2p with HAP2 domains of Cr, Lm, Tc, Tt, At, Tg, and Cm was performed and also revealed sequence conservation (Fig. 1D). A further sequence analysis of the HAP2 domain (PFAM10699) of different Plasmodium species revealed high conservation (Figure S1A). 3D structural models for PfHAP2 (Fig. 1E) and PfHAP2p (Fig. 1F) were predicted and generated using a Phyre2 [25]. Several sections of the PfHAP2 and PfHAP2p models exhibited conserved structural folds similar to viral class II fusion proteins, AtHAP2 (PDB ID: 5OW3) (Fig. 1G) and CrHAP2 (PDB ID:5MF1) (Fig. 1H). Despite the similarities, however, sections of the PfHAP2 and PfHAP2p models were comprised of domains which are dissimilar to AtHAP2 and CrHAP2. Comparison of PfHAP2 and PfHAP2p predicted structures with structures from At and Cr, revealed key differences in the cd-loop region. While the cd-loop regions in At (Fig. 1G) and Cr (Fig. 1H) were comprised of helical structures, the PfHAP2 cd-loop contained two antiparallel β strands (Figure S1B and Figure S1C) and the PfHAP2p cd-loop exhibited a flexible loop like structure (Figure S1D and Figure S1E). Key differences were also observed between the models of PfHAP2 and PfHAP2p (Fig. 1I) across all the domains, including domain I (Fig. 1J), domain II (Fig. 1K), cd-loop (Fig. 1L), and domain III (Fig. 1M). The domain II of HAP2p exhibited a 3-stranded antiparallel β sheet, which was completely absent in the HAP2 (Fig. 1K,L). Additionally, an α-helix of the Domain III from HAP2p was found to be replaced with a flexible loop like structure in HAP2 (Fig. 1M). While the C-terminal regions of HAP2 and HAP2p models also exhibited structural variations (Fig. 1N), an exact comparison was not made as most C-terminal residues of HAP2p could not be modelled.
Two HAP2/GCS1 genes are highly conserved in P. falciparum. A, B Domain architecture of two HAP2/GCS1 family proteins from P. falciparum indicating SP (signal peptide), in black; two discontinuous domains [I (in red) and II (in yellow)] followed by an immunoglobulin domain III (in blue), transmembrane (TM) domain in pink; cytoplasmic region in grey; cd-loop region and signature HAP2/PFAM10699 domain are marked separately. Amino acids indicate respective positions of various domains. C Part of domain II (DII) of HAP2 proteins from P. falciparum (PfHAP2 and PfHAP2p), Arabidopsis thaliana (At), Toxoplasma gondii (Tg), Eimeria. tenella (Et), Crytosporidium muris (Cm), Chlamydomonas reinhardtii (Cr), Tetrahymena thermophile (Tt), Leishmania major (Lm). Conserved cysteine residues are in white font on a red background, other conserved residues are in red font and marked in blue boxes, dots indicate indels present in various species. The orange line above the sequence alignment indicates the cd-loop of PfHAP2p. D HAP2/PFAM10699 domain of HAP2 proteins from P. falciparum (PfHAP2 and PfHAP2p), At, Tg, Et, Cm, Cr, Tt and Lm. Conserved residues are in white font on a red background. E, F Homology modeling predicted three-dimensional structures of PfHAP2 and PfHAP2p showing different domains. G, H Known three-dimensional structures of AtHAP2 (PDB: 5OW4) and CrHAP2 (PDB: CrHAP2) showing different domains (Domain I in red, Domain II in yellow, cd-loop in orange, Domain III in blue, C-terminal regions in grey). I Superimposed structures of PfHAP2 and PfHAP2p showing differences in various domains (PfHAP2: Domain I in red, Domain II in yellow, cd-loop in orange, Domain III in blue, C-terminal regions in grey; PfHAP2p: Domain I in cyan, Domain II in marine, cd-loop in chartreuse, Domain III in magenta, C-terminal regions in brown). Zoomed in view of the superimposed structures of PfHAP2 and PfHAP2p showing differences in domain I (J) (Red: PfHAP2, cyan: PfHAP2p), domain II (K) (Yellow: PfHAP2, marine: PfHAP2p), cd-loop (L) (Orange: PfHAP2, chartreuse: PfHAP2p), domain III (M) (Blue: PfHAP2, magenta: PfHAP2p) and c-terminal regions (N) (Grey: PfHAP2, brown: PfHAP2p). See also Figure S1
PfHAP2 and PfHAP2p are expressed in gametocytes and gametes
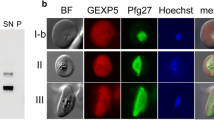
To examine expression and localization of PfHAP2 and PfHAP2p in the sexual stages, two separate transgenic parasite lines were engineered to express GFP-tagged PfHAP2 (PfHAP2GFP) and mCherry-tagged PfHAP2p (PfHAP2pmCherry), using double-crossover homologous recombination (Figure S2). In addition, antibodies were generated against PfHAP2p using a KLH-conjugated peptide representing the cd-fusion loop (KLH-RQDDMVFPLFSHFNYKKAKFKC). Genotyping PCRs were performed to confirm the successful generation of transgenic PfHAP2GFP parasites (Figure S2B and S2C) and transgenic PfHAP2pmCherry parasites (Figure S2D and S2E). We then performed dual indirect immunofluorescence assays (IFAs) using anti-GFP antibody and anti-P230p antibody (an antibody specific to stage V male gametocytes), as well as anti-GFP antibody and anti-Pfg377 antibody (an antibody specific to female gametocytes). This revealed that PfHAP2 was expressed internally in male stage V gametocytes while in the microgametes, it was expressed throughout the flagellum (Fig. 2A). PfHAP2 expression was also detected in stage II, III and IV male gametocytes (Figure S3A). PfHAP2 expression was not detected in female stage V gametocytes (Fig. 2B). For further PfHAP2p expression analysis, dual IFAs were performed using anti-mCherry and anti-tubulin antibody (an antibody specific to male stage V gametocytes and microgametes), as well as anti-mCherry antibody and anti-Pfg377 antibody. This revealed that PfHAP2p was not expressed in stage II and III gametocytes (data not shown) but was expressed internally in male stage V gametocytes and throughout the flagellum (Fig. 2C). Unexpectedly, it was also expressed in female stage V gametocytes (Fig. 2D). PfHAP2p expression was also detected in activated male gametocytes (Figure S3B) and activated female gametocytes (Figure S3C). PfHAP2GFP parasites and PfHAP2pmCherry parasites infected mosquitoes and formed oocysts similar to wildtype (WT) PfNF54 parasites, indicating that the addition of fluorescent tags to the C-termini of proteins did not interfere with their putative function (data not shown). These parasites were next used for creating a genetic cross (PfHAP2GFP × PfHAP2pmCherry) to examine expression and localization of PfHAP2 and PfHAP2p within the same parasite (Figure S4A). Genotyping PCRs were performed to confirm the successful generation of double transgenic PfHAP2GFP × PfHAP2pmCherry parasites (Figure S4B, S4C and S4D). Dual IFAs using anti-GFP antibody and anti-mCherry antibody on PfHAP2GFP × PfHAP2pmCherry gametocytes revealed that while PfHAP2 is expressed in stage II–stage V throughout the body of the gametocytes with maximum expression in stage V, HAP2p is expressed in stage IV–stage V with maximum expression in stage V gametocytes (Fig. 2F). HAP2 and HAP2p were both expressed in the same microgametes (Fig. 2F) and partially co-localized in the flagellum.
PfHAP2 and PfHAP2p are expressed in gametocytes and gametes. A, B IFAs were performed on PfHAP2GFP stage V gametocytes and 20 min post activation using anti-GFP (in red) and anti-PfP230p antibodies (in green) for male gametocytes and microgametes and using anti-GFP (in red) and anti-Pfg377 antibodies (in green) for female gametocytes. C, D IFAs were performed on PfHAP2pmCherry stage V gametocytes and microgametes using anti-mCherry (in red) and anti-P230p antibodies for male gametocytes, anti-Tubulin antibodies (in green) for microgametes and E using anti-mCherry (in red) and anti-Pfg377 antibodies (in green) for female gametocytes. F IFAs were performed on double transgenic PfHAP2GFP × PfHAP2pmCherry stage II–V gametocytes and microgametes using anti-GFP (in green) and anti-mCherry antibodies (in red)
PfHAP2 and PfHAP2p are each critical for fertilization
To determine the importance of PfHAP2 and PfHAP2p in fertilization, we created gene deletion parasites using a CRISPR/Cas9 strategy (Figure S5). Two clonal lines each for Pfhap2¯ and Pfhap2p¯ individual gene knockouts were isolated by limiting dilution and successful gene deletions were confirmed by genotyping PCR (Figure S5). For phenotypic analysis of Pfhap2¯ and Pfhap2p¯ parasite clones, we performed asexual blood stage growth assays. Intraerythrocytic development was compared to WT PfNF54 parasites over two replication cycles using Giemsa-stained culture smears. These assays showed that Pfhap2¯ and Pfhap2p¯ asexual stages did not exhibit any apparent growth defect (Fig. 3A). We next examined gametocyte formation and differentiation of Pfhap2¯ and Pfhap2p¯ parasites. No significant defect in gametocyte development and therefore, gametocytemia was observed on day 15 of gametocyte culture for Pfhap2¯ and Pfhap2p¯ when compared to gametocytemia for WT PfNF54 control (Fig. 3B). To analyze microgamete formation, we assessed exflagellation, the rapid process by which microgametes form within activated male gametocytes and egress from the infected RBCs. Pfhap2¯ and Pfhap2p¯ stage V gametocytes were activated by adding human serum and a temperature shift to room temperature (RT). There were no significant differences observed in the number of exflagellation centers of Pfhap2¯ and Pfhap2p¯ and WT PfNF54 control (Fig. 3C). Next, to ascertain the role of PfHAP2 and PfHAP2p in fertilization, we analyzed zygote formation 15 hours post gametocyte activation. Anti-Pfs25 antibody was used to stain WT PfNF54, Pfhap2¯ and Pfhap2p¯ activated cultures, which revealed no zygotes in Pfhap2¯ and Pfhap2p¯ parasite cultures. This stood in stark contrast to WT PfNF54 control, which showed normal zygote formation (Fig. 3D). These results indicate that both PfHAP2 and PfHAP2p are individually essential for zygote formation.
PfHAP2 and PfHAP2p are each essential for fertilization. A Ring stage synchronous cultures for WT PfNF54 and two different clones of Pfhap2¯ (clone E5 and F9) and Pfhap2p¯ (clone cl1 and cl3) were set up at 1% parasitemia and parasite growth was measured over the course of two erythrocytic infection cycles using Giemsa-stained culture smears. Data were averaged from three biological replicates and presented as the mean ± standard deviation (SD). B Ring stage synchronous cultures for WT PfNF54 and Pfhap2¯ (clone E5 and F9) and Pfhap2p¯ (clone 1 and clone 3) were set up at 1% parasitemia for in vitro generation of gametocytes and gametocytemia was measured on day 15 using Giemsa-stained smears. Data were averaged from three biological replicates and presented as the mean ± standard deviation (SD). C Number of exflagellation centers (emergence of microgametes) per field at 15 min post-activation. Data were averaged from three biological replicates and presented as the mean ± standard deviation (SD). D Zygotes were immunolabelled with anti-Pfs25 antibody and counted in 50 optical fields in triplicate. Results are shown as mean ± SD. ND-Not detected
PfHAP2 and PfHAP2p are each essential for fertility of male gametes
After ascertaining the critical roles of PfHAP2 and PfHAP2p in fertilization, we examined the transmissibility of Pfhap2¯ and Pfhap2p¯ parasites to the mosquito vector. For this, stage V gametocytes of Pfhap2¯, Pfhap2p¯ and WT PfNF54 parasites were mixed with human serum and human red blood cells (RBCs) to prepare infectious blood meals for Anopheles stephensi mosquitoes. Analysis of mosquito midgut infection 7 days post blood meal showed no oocysts in the Pfhap2¯ and Pfhap2p¯ lines, demonstrating that they are each required for parasite transmission to mosquitoes (Fig. 4A). We next examined a sex-specific function for PfHAP2 and PfHAP2p using genetic crosses with Pfhap2¯ and Pfhap2p¯ parasites in combination with transgenic parasite lines which formed either fertile female gametes only (Pfcdpk4¯) [26] or fertile male gametes only (Pfmacfet¯) [27]. WT PfNF54, Pfhap2¯, Pfhap2p¯, Pfcdpk4¯, Pfmacfet¯ gametocytes were generated in vitro and cultures were first individually fed to A. stephensi mosquitoes on day 15 of in vitro culture. Oocyst numbers in the mosquito midgut were quantified on day 7 post blood meal and revealed that, as expected, only the WT PfNF54 parasites infected mosquitoes and formed oocysts. For genetic crosses, gametocytes from the same cultures were mixed in pairs as follows: Pfhap2¯ × Pfcdpk4¯, Pfhap2p¯ × Pfcdpk4¯, Pfhap2¯ × Pfmacfet¯, Pfhap2p¯ × Pfmacfet¯, Pfcdpk4¯ × Pfmacfet¯. These mixed cultures were fed to mosquitoes and oocysts were enumerated in the mosquito midguts on day 7 post feeding for all the crosses (Fig. 4B). As expected, the Pfcdpk4¯ “female” × Pfmacfet¯ “male” cross showed robust oocyst numbers, comparable to WT PfNF54 control infections because the Pfmacfet¯ male gametes could fertilize Pfcdpk4¯ female gametes. In contrast, the Pfhap2¯ × Pfhap2p¯ cross showed no oocysts, indicating that both proteins are either essential either for the same sex or both sexes. The Pfhap2¯ × Pfmacfet¯ cross however showed robust oocyst numbers, indicating that the female Pfhap2¯ gametes were productively fertilized by male Pfmacfet¯ gametes (Fig. 4B). Conversely, in the Pfhap2¯ × Pfcdpk4¯ cross no oocysts were observed, indicating that male Pfhap2¯ gametes were unable to fertilize fertile female Pfcdpk4¯ gametes (Fig. 4B). These results show that Pfhap2¯ males are sterile but females are fertile. For the Pfhap2p¯ crosses, we observed that Pfhap2p¯ gametes were productively fertilized by Pfmacfet¯ male gametes, resulting in oocyst development in the mosquito midguts (Fig. 4B) but in the Pfhap2p¯ × Pfcdpk4¯ cross, male Pfhap2p¯ gametes were unable to fertilize female Pfcdpk4¯ gametes and thus, no oocyst development was observed (Fig. 4B). These results show that Pfhap2p¯ males are sterile but females are fertile. Together, these genetic crossing experiments demonstrate that both PfHAP2 and PfHAP2p individually have critical male-contributed roles in gamete fertility.
PfHAP2 and PfHAP2p are critical for zygote formation and have male sex restricted function. A Mosquitoes were dissected on day 7 post feed and number of oocysts were measured per midgut. Data were averaged from three biological replicates with a minimum of 50 mosquito guts and presented as the mean ± standard deviation (SD). ND not detected. B Oocyst formation of WT PfNF54, Pfhap2¯, Pfhap2p¯, Pfcdpk4¯, Pfmacfet¯, Pfhap2¯ × Pfcdpk4¯, Pfhap2p¯ × Pfcdpk4¯, Pfhap2¯ × Pfmacfet¯, Pfhap2p¯ × Pfmacfet¯, Pfcdpk4¯ × Pfmacfet¯, Pfhap2¯ × Pfhap2p¯. In vitro genetic crosses demonstrated that the Pfhap2¯ and Pfhap2p¯ showed productive cross-fertilization with the Pfmacfet¯ parasites (which produces functional males only), and not with Pfcdpk4¯ (which produces functional females only) (error bar indicates mean ± SD; n = 2)
PfHAP2 and PfHAP2p are functional cell fusogens
Arabidopsis thaliana (At) HAP2/GCS1 was shown to be fusogenic in cell–cell and virus-cell fusion assays [21]. To determine whether PfHAP2 and PfHAP2p are authentic membrane fusogens, we tested whether they could mediate fusion of mammalian Baby Hamster Kidney (BHK) cells. We synthesized the complete codon-optimized coding regions of PfHAP2 and PfHAP2p and cloned them independently into pCI plasmids containing IRES controlled reporters that enable the quantification of cell content mixing (see Materials and Methods). We also appended the signal peptide (SP) of PfHAP2 to the PfHAP2p N-terminus because the endogenous protein lacks an apparent secretion signal. To determine whether mammalian cells were able to express PfHAP2 and PfHAP2p, we also included V5 tags in the C-termini of the predicted proteins and performed immunofluorescence microscopy, demonstrating that both proteins were independently expressed (Figure S5). Each plasmid contained a different reporter gene encoding a fluorescent protein with a distinct location. To measure PfHAP2 cell fusion potential, we transfected BHK-cells either with pCI::PfHAP2::GFPnes (GFP in the cytoplasm) or pCI::PfHAP2::H2B-RFP (RFP in the nucleus) and mixed the two populations. 42 h post-mixing, we quantified hybrid cells containing mixed green cytoplasm with red nuclei (Fig. 5). To measure PfHAP2p cell fusion potential, we transfected BHK-cells with either pCI::PfHAP2p::GFPnes or with pCI::PfHAP2p::H2B-RFP and mixed the two populations. Following 42 h post-mixing, we again quantified hybrid cells containing mixed green cytoplasm with red nuclei (Fig. 5). We found that both PfHAP2 and PfHAP2p induced cytoplasmic content mixing (i.e., cell fusion) to similar levels as the AtHAP2 (Fig. 5A–C). Thus, both PfHAP2 and PfHAP2p can independently mediate cell fusion. To observe the cell fusion events as they occur in the culture, we performed time-lapse imaging using myristoylated GFP as a membrane marker co-expressed with PfHAP2 and imaged by spinning disk confocal microscopy. Figure 5D shows how two cells expressing PfHAP2 recognized each other and fused to merge their cytoplasms revealing cell–cell fusion (Movie S1).
PfHAP2 and PfHAP2p mediate cell–cell fusion in mammalian cells. A–C BHK-21 cell–cell fusion was measured by content-mixing, indicated by the appearance of multinucleated cells containing blue nuclei (DAPI), magenta nuclei (H2B-RFP) and green cytoplasm (GFPnes). A Representative images of mixed cells. Scale bars 20 µm. B Cartoon of experimental design. C Quantification of content mixing. Cell–cell fusion was measured by content mixing, indicated by the presence of multinucleated cells containing magenta (H2B-RFP) or/and DAPI nuclei within cells with GFP cytoplasm (GFPnes). The mixing indices presented as means ± SEM of at least three independent trials. Comparisons by one way ANOVA followed by Bonferroni’s test. **p < 0.01; ***p < 0.0005; ****p < 0.0001. D Spinning disk confocal microscopy time-lapse images indicating the merging of two BHK cells expressing myristoylated-GFP (myrGFP) and PfHAP2. Time in hours:minutes. The green channel (GFP, white) and a merge of the DIC (grey) and Hoechst (magenta) are shown. White arrows and arrowheads indicate the fusing and fused cells, respectively. Scale bar 20 µm. (Refer to Supplementary Movie 1)
P. falciparum HAP2 and HAP2p fuse BHK cells when homotypically expressed in both fusing cells
Cell fusion is a process that can be driven by a unidirectional mechanism in which only one of the fusing cells expresses the fusogenic protein, as is the case for known fusogens from enveloped viruses [28]. An alternative bidirectional mechanism requires the expression of the fusogens in both fusing membranes and this is characteristic of homotypic fusogens involved in fusion between somatic cells, for example, mediated by the EFF-1 or AFF-1 proteins from C. elegans [28,29,30,31,32,33] and Myomaker in vertebrate muscles [30]. In addition, the intracellular SNARE proteins are required to dock and fuse membranes and they must be present in both interacting membranes [36]. Our evidence from genetic crosses indicated that both PfHAP2 and PfHAP2p are male-contributed factors each critical for fertilization. To test whether PfHAP2 and PfHAP2p work unidirectionally or bidirectionally, we transfected BHK cells with PfHAP2 or PfHAP2p together with a cytoplasmic GFP and mixed them with cells expressing only the vector encoding a nuclear H2B::RFP. Presence of multinucleated cells with only green cytoplasm would reveal fusion between cells expressing the fusogens; in contrast, the appearance of mixed cells with green cytoplasm and red nuclei would reveal unilateral fusion. As negative control we used BHK-GFPnes cells mixed with BHK-H2B-RFP and we found mostly mononucleated cells and some binucleated dividing cells expressing only green cytoplasm (GFPnes) or cells expressing only red nuclei (H2B-RFP). We found that both PfHAP2 and PfHAP2p expressing cells showed multinucleated cells expressing 2–8 nuclei with green cytoplasm (Fig. 6A, B). Similarly, AtHAP2 showed a similar result with 20–40% multinucleation (Fig. 6A, B). As a unilateral positive control, we used the Vesicular Stomatitis Virus-G glycoprotein and found an average of 80% multinucleation at neutral pH and 100% multinucleation at the optimal low pH. While the VSV-G expressing cells showed 50% multinucleated cells with mixed red nuclei and green cytoplasm at neutral pH and 100% after incubation at low pH, PfHAP2, PfHAP2p and AtHAP2 showed very few cells with mixed cytoplasms at levels similar to the negative control cells expressing only RFP or GFP (Fig. 6C). Moreover, expression of PfHAP2 and PfHAP2p in trans (different cells) or in cis (in the same cells) was unable to fuse cells and no mixing was observed (Fig. 6A, C). Thus, the P. falciparum HAP2 fusogens failed to fuse heterologous mammalian BHK cells in a unidirectional (viral-like) fashion. The proteins must be present in both fusing cells.
PfHAP2 and PfHAP2p mediated cell–cell fusion is bidirectional in mammalian cells. Unidirectional BHK-21 cell–cell fusion was measured by content-mixing, indicated by the presence of multinucleated cells containing magenta nuclei (H2B-RFP) and green cytoplasm (GFPnes). A Images of mixed cells. Unilateral fusion was evaluated by mixing control cells expressing nuclear H2B-RFP (magenta) with cells expressing GFP with a nuclear export signal (GFPnes, green cytoplasm) only or together with AtHAP2, PfHAP2, PfHAP2p, PfHAP2 and PfHAP2p (in cis) or VSV G incubated at neutral or low pH. Multinucleated cells labeled with DAPI only (arrowheads) or with both markers (arrows; green cytoplasm and magenta nuclei). Scale bars 20 µm. B Multinucleation index. C Quantification of content-mixing experiments. Bar chart showing means ± SEM of four independent experiments. Comparisons by one-way ANOVA followed by Dunett’s test against the vector. ns non-significant, **p < 0.01, ****p < 0.0001.
Discussion
Fertilization is the central event during sexual reproduction when two gametes interact, and their plasma membranes fuse to form a zygote. This process generates an incipient genetically unique organism, creating genetic diversity in a population. The fusion of Plasmodium gametes during fertilization is an essential step in parasite transmission and occurs inside the mosquito vector. It creates novel parasite strains, some of them poised to have selective fitness advantages over their parents. However, the molecular mechanisms leading to fertilization are poorly understood in Plasmodium. Herein, we demonstrate that the human malaria parasite P. falciparum expresses two HAP2/GCS1 family members, PfHAP2 and PfHAP2p and present genetic evidence that each is critical for male fertility only. We further demonstrate that PfHAP2 and PfHAP2p can independently mediate cell membrane fusion and thus are authentic cell/gamete fusogens (Fig. 7).
Model for PfHAP2 and PfHAP2p function during Plasmodium gamete fusion. PfHAP2 and PfHAP2p are expressed in male microgametes while PfHAP2p is also expressed in female macrogametes. The evidence from genetic crosses shows that both proteins are male-only contributed factors that are each essential for fertilization. This suggests that they are deployed by the microgamete to mediate membrane fusion with the macrogamete and that their mode of action is unidirectional (top panel). However, evidence from the cell fusion experiments shows that each PfHAP2 and PfHAP2p must be present on both cells that engage in fusion (bottom panel) and that this bidirectional requirement is homotypic. PfHAP2 and PfHAP2p might form dimeric or trimeric complexes during gamete fusion as has been shown for other membrane fusogens. This figure was created using Biorender
HAP2/GCS1 family proteins are transmembrane proteins with large extracellular regions containing three globular domains [19–21]. HAP2 was initially identified in the flowering plant A. thaliana [17, 34] via a mutagenesis screen for male sterile lines, and in Lilium longiflorum pollen [18] as a sperm-specific protein involved in sperm-egg fusion. HAP2 proteins are the only gamete fusogens present in three out of four eukaryotic kingdoms [19, 35,36,37]. They share similarity with class II viral fusion proteins [19–21]. One proposed model of action is that upon a trigger, a short conserved hydrophobic region (cd-loop) of HAP2 proteins becomes exposed on the male gamete plasma membrane and inserts into the female gamete plasma membrane while forming trimers, thereby overcoming the energetic barriers that are normally preventing membrane fusion [19, 20, 35]. This viral-like unilateral mechanism of action is different from the bilateral mechanism proposed for the related C. elegans EFF-1 and AFF-1. These somatic cell fusogens, structurally and functionally related to HAP2, act in the respective organism and in heterologous systems on both fusing membranes [28,29,30,31,32,33]. Interestingly, the AtHAP2 also mediates bilateral cell fusion in BHK cells and in virus-cell fusion assays [21].
Previous studies identified a HAP2 ortholog in the rodent malaria parasite P. berghei and showed that PbHAP2 functions in fertilization [22, 24], however, its precise molecular function remained undefined. We show here that Plasmodium species possess two HAP2/GCS1 family proteins and demonstrate that they are authentic fusogens in a heterologous cell expression system and each is essential for fertilization in the human malaria parasite P. falciparum. So far, only Dictyostelium discoideum was shown to encode two paralogs of HAP2 related proteins, which are both critical for mating [38]. We found that PfHAP2 is expressed during male gametocytogenesis and in male microgametes, while PfHAP2p was only detected in late-stage V gametocytes and microgametes but was also expressed in female gametocytes and macrogametes. Yet, in IFAs performed on live, non-permeabilized activated gametocytes/gametes, we did not observe staining with anti-PfHAP2p antiserum. This suggests that PfHAP2p and possibly PfHAP2 are translocated to the plasma membrane and localize on the surface just-in-time before gamete fusion. Gene deletions for HAP2 and HAP2p showed that neither of these proteins are essential for gametocyte formation and maturation. The formation and emergence (exflagellation) of microgametes was also unaffected in both Pfhap2¯ and Pfhap2p¯ parasites but importantly, these gene deletion strains did not infect mosquitoes. Further analysis revealed that zygote formation was completely abolished in Pfhap2¯ and Pfhap2p¯ parasites, establishing functions for these proteins in fertilization and explaining the complete defect in transmission. Genetic crosses of these parasites with sex-specific sterile transgenic parasites demonstrated that both PfHAP2 and PfHAP2p are independently contributed by microgametes as essential for fertility but are not contributed for fertility by macrogametes.
Using codon-optimized, full-length constructs, we successfully expressed PfHAP2 and PfHAP2p in the mammalian BHK cell line and used these cells in cell fusion assays [21]. To achieve this for PfHAP2p however, we appended a signal peptide (SP) to its N-terminus because an apparent secretion signal is lacking in the endogenous protein. Using this assay, we demonstrated that both PfHAP2 and PfHAP2p possess independent fusogenic activity as their expression mediated cell fusion, cell content mixing, and syncytia formation. The magnitude of PfHAP2 and PfHAP2p cell fusion activity in this assay was comparable to the activity of AtHAP2, indicating that the Plasmodium proteins are potent membrane fusogens. Our genetic crosses clearly demonstrated a requirement for PfHAP2 and PfHAP2p in the microgametes only. It is thus reasonable to assume that these fusogens are utilized unilaterally by the male to mediate fusion with the female (Fig. 7). In contrast however, the results in heterologous BHK cell fusion assays showed that in this in vitro system, the proteins cannot mediate unidirectional cell–cell fusion. Only the expression of either PfHAP2 or PfHAP2p in both fusing cells mediated bidirectional cell–cell fusion (Fig. 7). It is of interest to point out that similar differences between genetic and cell fusion data with regard to directionality of the fusion event has also been observed for the AtHAP [21]. These contradictory lines of evidence might be reconciled by the possibility that in the parasite, additional proteins of unknown identity on the female interact with HAP2 and HAP2p on the male, to enable unidirectional gamete fusion activity. The absence of such proteins in the BHK cells might, however, require HAP2 or HAP2p to be present on both fusing cells. Thus, the genetic and cell fusion data require conciliation with future experimentation. Because PfHAP2p did not exhibit a predicted canonical SP that could mediate its entry into the parasite’s secretory pathway, it might require an escort partner protein. Alternatively, PfHAP2p might contain a non-conventional SP that was not identifiable by bioinformatic prediction, or this protein might be trafficked to the parasite plasma membrane by an unconventional secretion mechanism described in Plasmodium and other protozoans [39]. Once positioned on the microgamete plasma membrane, PfHAP2 and PfHAP2p might form trimers (Fig. 7) as has been shown for HAP2 proteins of other organisms. To understand potential complex formation of PfHAP2 and PfHAP2p will require direct future biochemical characterization in microgametes.
The domain organization of PfHAP2 and PfHA2p is similar with an extracellular domain comprising domains I, II and III followed by a stem, membrane proximal region and transmembrane domain, which are typically present in HAP2/GCS1 family proteins. However, PfHAP2 and PfHAP2p show predicted structural differences across domain I, domain II, and domain III. Intriguingly, different conformations were observed between the cd-loops of PfHAP2 and PfHAP2p. HAP2 might have evolved multiple modes of membrane insertion to accommodate the diversity of membrane environments it has encountered during eukaryotic evolution [35]. Previously, cd-fusion loop differences of Arabidopsis, Trypanosoma and Chlamydomonas HAP2 have been revealed by crystal structures [35]. It is also possible that different HAP2 cd-loops have differential lipid binding affinities towards different target membrane lipid compositions, and it will be very interesting to determine if PfHAP2 and PfHAP2p have differential membrane lipid binding affinities in future studies. The recent discovery of HAP2-related Fusexin1 proteins in integrated mobile elements in archaea suggests novel mechanisms of horizontal gene transfer [40] that might help to explain the evolution of the fusexin family in eukaryotes, prokaryotes and the virome.
The transmission of sexual stages from the human host to the mosquito vector represents a major bottleneck in the malaria parasite life cycle [41]. This has been identified as a vulnerability in transmission and transmission blocking vaccines targeting surface molecules in gametocytes, gametes, zygotes and ookinetes have been proposed [42, 43]. Previous work has shown that immunizations with recombinant protein fragments encompassing amino acids 355–609 of PbHAP2 and 195–684 of PfHAP2 generated transmission blocking activity [44, 45]. Furthermore, antibodies targeting the cd-loop of PbHAP2 and PfHAP2 had shown transmission blocking activity with a 58.9% and 75.5% reduction in mosquito infection, respectively [46]. Very recently, antibodies targeting the domain III of PbHAP2 have also been shown to block fertilization in vitro and transmission to mosquitoes [47]. Thus, future studies could focus on identifying and targeting conserved antibody epitopes between PfHAP2 and PfHAP2p that could be simultaneously targeted to disrupt their respective fusogenic function.
In conclusion, we have shown herein that malaria parasites utilize two distinct HAP proteins, HAP2 and HAP2p, which can each independently mediate plasma membrane fusion. Each of these fusogens alone is critical for fertilization, and it remains to be determined whether they operate unidirectionally or bidirectionally in Plasmodium gamete fusion, together in a complex or serve individual and distinct functions. Furthermore, the signaling events that lead to the just-in-time surface-positioning and activation of PfHAP2 and PfHAP2p remain unknown and require future investigation.
Methods
Reagents and primary antibodies
All molecular biology reagents and oligonucleotides were purchased from Sigma–Aldrich. The following primary antibodies and antisera were used: mouse anti-GFP antibody (1:500, abcam, cat# ab1218 and cat# ab290); mouse anti-V5 antibody (1:500, Invitrogen, cat# R960-25); mouse anti-α-tubulin antibody (1:250, Sigma–Aldrich, cat# T9026); mouse anti-Pfs25 (1:1, ATCC, cat# MRA-28), rat anti-mCherry clone 16D7 (1:100, Thermo Scientific cat# M11217); rabbit anti-Pfg377 (1:500, kindly gifted by Professor Pietro Alano at Istituto Superiore di Sanità, Italy). The generation of polyclonal rabbit antisera against PfHAP2p (1:100) is described below.
Sequence analysis and structure predictions
Plasmodium DNA and protein sequences were retrieved from PlasmoDB (http://plasmodb.org/plasmo/) and aligned using Multalign (http://multalin.toulouse.inra.fr/multalin/) and Espript 3 (http://espript.ibcp.fr/ESPript/ESPript/) programs. Signal peptide, transmembrane domains, and secondary structures were predicted using the SignalP-5.0 Server, then TMHMM Server v.2.0, and Phyre2 respectively. The models for P. falciparum HAP2 (all residues) and HAP2p (residues 1–515) were generated using the Phyre2 Server. The templates utilized by the Phyre2 intensive search algorithm were A. thaliana (PDB ID-5OW3), C. reinhardtii (PDB ID-5MF1) and T. cruzi (PDB ID-5OW4). For PfHAP2, 488 (55%) residues modelled at greater than 90% confidence, using A. thaliana (20% identity), C. reinhardtii (21% identity) and T. cruzi (25% identity) structures as the templates. For PfHAP2p, 409 (41%) residues modelled at greater than 90% confidence using re from T. cruzi (23% identity), C. reinhardtii (17% identity) and A. thaliana (19% identity) structures as the templates. The models generated were analyzed using ORION server to identify the fold and structurally similar molecules from the PDB. All structural comparisons and visualizations were performed using Pymol 2.0.
Plasmodium falciparum strains and culturing
P. falciparum NF54 was cultured as asexual blood stages according to standard procedures [48] and received medium changes every 24 h. Gametocytes were generated using O+ human RBCs (Valley Biomedical, VA, US) and O+ human serum (Interstate Blood Bank, TN, US) using previously published protocols [49, 50]. Gametocyte cultures were set up either in T150 cm2 flasks containing a final volume of 50 mL at 1% starting parasitemia and 5% hematocrit or in 6 well plates with a final volume of 5 mL at 1% starting parasitemia and 4% hematocrit. Cultures were supplied with complete RPMI medium supplemented with 0.05 g/L hypoxanthine, 0.3 mg/L l-glutamine and 10% (v/v) human serum. Cultures were kept at 37 °C and supplemented with gas containing 3% O2/5% CO2/92% N2.
Generation of anti-PfHAP2p antibodies
Peptide corresponding to amino acids 95–116 of PfHAP2P (RQDDMVFPLFSHFNYKKAKFKC), containing a fragment of the cd fusion loop, conjugated to carrier protein Keyhole limpet hemocyanin (KLH)—was synthesized by Biomatic Inc. (DE, US) and were used for immunization of animals. PfHAP2p antisera was generated by immunizing rabbits by Antagene Inc. (CA, US) and prescribes guidelines for animal handling were followed. IgG purification and ELISA was performed by Antagene Inc.
Generation of Pfhap2¯, Pfhap2p¯and the creation of transgenic parasites expressing PfHAP2GFP and PfHAP2pmCherry
Oligonucleotide primers used for the creation and analysis of P. falciparum hap2¯, P. falciparum hap2p¯, PfHAP2GFP and PfHAP2mCherry parasites are detailed in Table S1. Deletion of PfHAP2 (PlasmoDB identifier PF3D7_1014200) and PfHAP2p (PlasmoDB identifier PF3D7_0816300) was achieved based on the previously reported CRISPR/Cas9 strategy using pYC plasmid [51]. PfHAP2 and PfHAP2p were deleted using double crossover homologous recombination. Complementary regions of PfHAP2 or PfHAP2p upstream and downstream of the open reading frame were ligated into plasmid pFCL3 (generated by modification in pYC plasmid and described in [52] as was the 20-nucleotide guide RNA sequence [51] resulting in the creation of a plasmid pFCL3_HAP2_KO and pFCL3_HAP2p_KO, respectively. Approximately 100 µg plasmid DNA was transfected into the P. falciparum NF54 strain using a previously published method [53] and selected using 8 nM WR99210 (gifted by Jacobus Pharmaceuticals). Gene deletion was confirmed by genotyping PCR (Fig. S5). Creation of PfHAP2-GFP and PfHAP2p-mCherry also used pFCL3, through the addition of a GFP epitope tag to the carboxy terminus of PfHAP2 and addition of a mCherry epitope tag to the carboxy terminus of PfHAP2p (Fig. S2). Two individual clones for Pfhap2¯, and Pfhap2p¯, and one clone each for PfHAP2GFP and PfHAP2mCherry were used for phenotypic analysis.
Generation of double transgenic PfHAP2GFP × PfHAP2pmCherrys parasites
A genetic cross was generated between PfHAP2GFP and PfHAP2pmCherrys parasites using FRG NOD huHep mice [54] with human chimeric livers and A. stephensi mosquitoes as described elsewhere [55]. The gametocytes for PfHAP2GFP and PfHAP2mCherry were cultured in 6-well plates and were mixed in equal ratio and fed to A. stephensi mosquitoes where gametes for these two parasites would fuse and form hybrid progeny along with selfed parasites (Figure S4A). The salivary gland sporozoites from day 14 infected mosquitoes, which would contain double transgenic PfHAP2GFP × PfHAP2pmCherrys hybrid parasites, were injected intravenously (IV), and parasites were transitioned to in vitro culture day 7 post injection. Parasites were cloned using limiting dilution and PfHAP2GFP × PfHAP2pmCherrys clonal parasites were confirmed by genotyping (Figure S4B, S4C and S4D). Clone C3 was used for performing IFAs for relative HAP2 and HAP2p expression analysis in the same parasite.
Measurement of asexual blood stage growth and gametocyte development
To compare asexual blood stage replication and growth between the WT PfNF54, Pfhap2¯ and Pfhap2p¯ parasites, synchronized parasites were set up at an initial parasitemia of 1% at ring stages and cultured in 6-well plates as described above. Parasites were removed at 48 and 96 h. for preparation of Giemsa smears and parasitemia was scored per 1000 erythrocytes.
To compare gametocyte formation for WT PfNF54, Pfhap2¯, Pfhap2p¯ gametocytes were cultured as described above. Parasites were removed on day 15 of in vitro culture for preparation of Giemsa smears and gametocytemia was scored per 1000 erythrocytes.
Measuring of exflagellation, standard membrane feeding assay (SMFA)
Gametocytes of WT PfNF54, Pfhap2¯ and Pfhap2p¯ were cultured as described above. Mosquitoes were reared and maintained on sugar water at 24 °C and 70% humidity. Gametocytes from all the cultures were analyzed for prevalence of stage V gametocytes using Giemsa smears. For assaying comparative exflagellation, equal volume of gametocytes from WT, Pfhap2¯ and Pfhap2p¯ were mixed with human serum and O+ RBCs (50:50) % (v/v) and incubated at room temperature for 10 min. Exflagellation was scored for WT PfNF54, Pfhap2¯ and Pfhap2p¯ parasites via light microscopy by counting exflagellation centers in 10 random optical fields of view at 40 × magnification in a bright field microscope.
For SMFA, stage V gametocytes were mixed with human serum and O+ RBCs mixture (50:50) % (v/v) to achieve a final gametocytemia of 0.5% and loaded on standard mosquito feeders. Mosquitoes were allowed to feed for approximately 25 min. Mosquitoes were dissected day 7 post-feeding and midguts were examined for oocysts by light microscopy.
Comparative zygote formation assay
For the analysis of zygote formation, mature day 15 WT, Pfhap2¯ and Pfhap2p¯ gametocytes were mixed with human serum and diluted to a final gametocytemia of 2%. Gametocyte cultures were activated in vitro by the addition of 100 μM xanthurenic acid followed by incubation for 30 min (macrogamete development) or 6 h. (zygote development) at room temperature (RT). An equal volume from each sample was loaded onto Teflon coated slides and Immunofluorescence assay (described below) was performed with an anti-Pfs25 antibody. Macrogametes or zygotes for each sample were counted microscopically in a total number of 1000 of RBCs.
Immunofluorescence assay (IFA) on Plasmodium falciparum parasites
For IFAs of gametocytes and exflagellating gametes, cultures were fixed in 4% paraformaldehyde/0.0025% glutaraldehyde in PBS in a microcentrifuge tube by end-over-end rotation for 30 min. For each incubation step during IFA on gametocytes microcentrifuge tubes were kept on end-over-end rotation. For IFAs on zygotes, smears were prepared on Teflon coated slides and fixed with 4% paraformaldehyde/0.0025% glutaraldehyde solution for 30 min. Slides were kept in a humidity chamber for each step. Fixed parasites were washed twice with PBS and permeabilized using 0.1% Triton X-100/PBS solution for 10 min. Parasites were washed with PBS and blocked with 3%BSA/PBS for 45 min. Primary antiserum in 3%BSA/PBS was added to the parasites and slides or microcentrifuge tubes were incubated at 4 °C. Following three washes of 5 min each with PBS, appropriate secondary antibodies (Alex Fluor-488/594, Molecular Probes) were used at 1:500 dilution along with DAPI for 45 min at RT. Parasites were washed five times before mounting in ProLong Antifade (Thermo Fisher Scientific) mounting reagent. Images were obtained using a 100 × 1.4 NA objective 90 (Olympus) on a DeltaVision Elite High-Resolution Microscope (GE Healthcare Life Sciences).
Mammalian cells culture
BHK-21 cells (kindly obtained from Judith White, University of Virginia) were maintained in DMEM supplemented with 10% FBS (Biological Industries), 100 U/mL penicillin, 100 µg/mL streptomycin (Biological Industries), 2 mM l-glutamine (Biological Industries), 1 mM sodium pyruvate (Gibco), and 30 mM HEPES buffer, pH 7.3, at 37 °C with 5% CO2. Transfections were performed using jetPRIME (Polyplus) according to the manufacturer’s instructions.
PfHAP2 and PfHAP2p DNA constructs for expression in mammalian cells
Full-length genes of PfHAP2 and PfHAP2p were synthesized after codon optimization for mammalian systems by IDT Inc. Since PfHAP2p does not have a predicted signal peptide, signal peptide of PfHAP2 was added at 5’ of codon optimized PfHAP2p sequence. These genes were then subcloned into corresponding backbones pCI::H2B-RFP and pCI::GFPnes which has a nuclear export signal (NES) at the C-terminus of GFP. The V5 tag was introduced at the C-terminus of PfHAP2 and PfHAP2p for immunofluorescence assays. See Supplementary Tables 1 and 2 for description of the plasmids and primers used.
Plasmodium proteins expression in mammalian cells and immunofluorescence
BHK-21 cells grown on 24-well tissue-culture plates with glass bottom or coverslips, were transfected when they reached 70% confluence using 1 µg pCI::PfHAP2::H2B-RFP or pCI:: PfHAP2p::H2B-RFP. 18 h post-transfection, 20 µM 5-fluoro-2ʹ-deoxyuridine (FdUrd) was added to the plates to block the cell cycle and 24 h later, the cells were fixed with 4% paraformaldehyde (PFA; EM grade, Bar Naor, Israel) in PBS, followed by incubation in 40 mM NH4Cl to block free aldehydes, permeabilized in 0.1% Triton X-100 and blocked in 1% Fetal Bovine Serum (FBS). We stained the cells with anti-V5 mAb (Invitrogen, 1:500), the secondary antibody was donkey anti–mouse coupled to Alexa Fluor 488 (Invitrogen, 1:500) and supplemented with 1 µg/mL DAPI [21]. Micrographs were obtained using wide-field illumination using an ELYRA system S.1 microscope and a Plan-Apochromat 63X NA 1.4 objective (Zeiss); images were recorded with an iXon + EMCCD camera (Andor).
Cell fusion assay in mammalian cells
BHK-21 cells at 70% confluence were transfected with 1 µg pCI::AtHAP2::GFPnes, pCI::AtHAP2::H2B-RFP, pCI::PfHAP2::GFPnes, pCI::PfHAP2::H2B-RFP, pCI::PfHAP2p::GFPnes and pCI::PfHAP2p::H2B-RFP respectively. Control cells were transfected with pCI::GFPnes or pCI::H2B-RFP (vectors). Four hours after transfection, the cells were washed 4 times with DMEM with 10% serum (Invitrogen), 4 times with PBS, and detached using Trypsin (Biological Industries). The transfected cells were collected in Eppendorf tubes, resuspended in DMEM 10% FBS, and counted. Equal amounts of H2B-RFP and GFPnes cells were mixed and seeded in glass-bottom plates (12-well black, glass-bottom #1.5H; Cellvis) and incubated at 37 °C in 5% CO2. 18 h after mixing 20 µM FdUrd was added to the plates and 24 h later, the cells were fixed with 4% PFA in PBS. Nuclei were stained with 1 µg/mL DAPI. Micrographs were obtained using wide-field illumination using an ELYRA system S.1 microscope and a Plan-Apochromat 20X NA 0.8 lens (Zeiss); images were recorded with a iXon + EMCCD camera (Andor). The GFP + RFP mixing index was calculated as the number of nuclei in mixed cells, GFPnes cytoplasm with red (H2B-RFP) and blue (DAPI) nuclei out of the total number of nuclei of fluorescent cells in contact and expressed as percentage.
Live imaging of BHK cells expressing PfHAP2
BHK-21 cells were plated on 15 mm glass bottom plates (Wuxi NEST Biotechnology Co., Ltd.) and transfected with 1 µg pCI:: PfHAP2::H2B-RFP together with 0.5 µg myristoylated-GFP (myr-GFP) [56]. 18 h after transfection, the nuclei were stained with 2 μg/mL Hoechst dye for 10 min at 37 °C and the cells washed once with fresh medium. Time-lapse microscopy at 33 °C and with 0.5% CO2 was performed to identify fusing cells using a spinning disc confocal microscope (CSU-X; Yokogawa Electric Corporation) with an Eclipse Ti and a Plan-Apochromat 20X (NA, 0.75; Nikon) objective. Images in differential interference contrast, DAPI and green channels were recorded every 4 min in different positions of the plate using high gain and minimum laser exposure. Time lapse images were captured with an iXon 3 EMCCD camera (Andor Technology). Image analyses were performed in MetaMorph (v7.8) (Molecular Devices) and ImageJ (v1.53c) (National Institutes of Health).
Cytoplasmic content mixing to determine unidirectional versus bidirectional mechanism
BHK-21 cells were transfected (as explained above) with 1 µg of each plasmid: pCI::GFPnes; pCI::AtHAP2- V5::GFPnes; pCI PfHAP2-V5::GFPnes; pCI::PfHAP2p::GFPnes; pCI::VSV-G::GFPnes; in 35 mm plates. Four hours after transfection, the cells were washed, detached, counted, and mixed with an equal number of pCI::H2B-RFP (empty vector) expressing cells, and mixed cells were incubated as previously described. In all cases, 18 h after mixing, 20 µM FdUrd was added to the plates, and 24 h later, the cells were fixed with 4% paraformaldehyde diluted in PBS (without calcium and magnesium). Nuclei were stained with 1 µg/mL DAPI. Images were obtained using wide-field illumination with an ELYRA system S.1 microscope as described above. The GFP + RFP mixing index was calculated as the number of nuclei in mixed cells, green cytoplasm (GFPnes) with red (H2B-RFP) and blue (DAPI) nuclei out of the total number of nuclei in fluorescent cells in contact. The multinucleation indexes were defined as the ratio between the number of nuclei in multinucleated cells (Nm) and the total number of nuclei in mononucleated cells and expressing cells that were in contact (Nc) but did not fuse, using the following equation: % multinucleation = Nm/(Nc + Nm) × 100. For pCI:: VSV-G::GFPnes mixed with pCI::RFP at low pH, 2 h before fixing DMEM medium was changed by HMS buffer pH5.5, incubated for 5 min, and changed again by DMEM and incubated for additional 2 h.
Cell fusion assay by content mixing to determine unidirectional versus bidirectional mechanism when both proteins are expressed in the same cell
BHK-21 cells were co-transfected with 1 µg pCI PfHAP2-V5::GFPnes and 1 µg pCI::PfHAP2p::GFPnes in 35 mm plates. Four hours after transfection, the cells were washed, detached, counted, and mixed with an equal number of pCI::H2B-RFP (empty vector) expressing cells; a mixing experiment was performed as described before.
Cell fusion assay by content mixing for interaction in trans
BHK-21 cells were transfected (as explained above) with 1 µg pCI::PfHAP2::GFPnes or pCI::PfHAP2p::RFP in 35 mm plates. Four hours after transfection, the cells were washed, detached, counted, and mixed, and the GFP + RFP mixing index was calculated as described before.
Statistical analysis
All data are expressed as mean ± SD. Statistical differences were determined using one-way ANOVA with post hoc Bonferroni multiple comparison test or unpaired two-tailed Student's t test, as indicated in the figure legends. Values of p < 0.05 were considered statistically significant. Significances were calculated using GraphPad Prism 8 and are represented in the Figures as follows: ns, not significant, p > 0.05; *p < 0.05; **p < 0.01; ***p < 0.001.
Data availability
All data are presented in the manuscript. Derived data supporting the findings of this study are available from the corresponding author on request.
References
de Jong RM et al (2020) Immunity against sexual stage Plasmodium falciparum and Plasmodium vivax parasites. Immunol Rev 293(1):190–215
Andreadaki M et al (2018) Sequential membrane rupture and vesiculation during Plasmodium berghei gametocyte egress from the red blood cell. Sci Rep 8(1):3543
Sinden RE et al (1978) Gametocyte and gamete development in Plasmodium falciparum. Proc R Soc Lond B Biol Sci 201(1145):375–399
Wirth CC, Pradel G (2012) Molecular mechanisms of host cell egress by malaria parasites. Int J Med Microbiol 302(4–5):172–178
Beier JC (1998) Malaria parasite development in mosquitoes. Annu Rev Entomol 43:519–543
Janse CJ et al (1986) Rapid repeated DNA replication during microgametogenesis and DNA synthesis in young zygotes of Plasmodium berghei. Trans R Soc Trop Med Hyg 80(1):154–157
Rosenberg R, Rungsiwongse J (1991) The number of sporozoites produced by individual malaria oocysts. Am J Trop Med Hyg 45(5):574–577
Eksi S et al (2006) Malaria transmission-blocking antigen, Pfs230, mediates human red blood cell binding to exflagellating male parasites and oocyst production. Mol Microbiol 61(4):991–998
Marin-Mogollon C et al (2018) The Plasmodium falciparum male gametocyte protein P230p, a paralog of P230, is vital for ookinete formation and mosquito transmission. Sci Rep 8(1):14902
van Dijk MR et al (2001) A central role for P48/45 in malaria parasite male gamete fertility. Cell 104(1):153–164
van Schaijk BC et al (2006) Pfs47, paralog of the male fertility factor Pfs48/45, is a female specific surface protein in Plasmodium falciparum. Mol Biochem Parasitol 149(2):216–222
van Dijk MR et al (2010) Three members of the 6-cys protein family of Plasmodium play a role in gamete fertility. PLoS Pathog 6(4):e1000853
Bianchi E, Wright GJ (2020) Find and fuse: unsolved mysteries in sperm-egg recognition. PLoS Biol 18(11):e3000953
Brukman NG et al (2019) How cells fuse. J Cell Biol 218(5):1436–1451
Cyprys P, Lindemeier M, Sprunck S (2019) Gamete fusion is facilitated by two sperm cell-expressed DUF679 membrane proteins. Nat Plants 5(3):253–257
Takahashi T et al (2018) The male gamete membrane protein DMP9/DAU2 is required for double fertilization in flowering plants. Development 145(23):dev170076
Johnson MA et al (2004) Arabidopsis hapless mutations define essential gametophytic functions. Genetics 168(2):971–982
Mori T et al (2006) GENERATIVE CELL SPECIFIC 1 is essential for angiosperm fertilization. Nat Cell Biol 8(1):64–71
Fedry J et al (2017) The ancient gamete fusogen HAP2 is a eukaryotic class II fusion protein. Cell 168(5):904-915.e10
Pinello JF et al (2017) Structure-function studies link class II viral fusogens with the ancestral gamete fusion protein HAP2. Curr Biol 27(5):651–660
Valansi C et al (2017) Arabidopsis HAP2/GCS1 is a gamete fusion protein homologous to somatic and viral fusogens. J Cell Biol 216(3):571–581
Liu Y et al (2008) The conserved plant sterility gene HAP2 functions after attachment of fusogenic membranes in Chlamydomonas and Plasmodium gametes. Genes Dev 22(8):1051–1068
Ramakrishnan C et al (2019) An experimental genetically attenuated live vaccine to prevent transmission of Toxoplasma gondii by cats. Sci Rep 9(1):1474
Hirai M et al (2008) Male fertility of malaria parasites is determined by GCS1, a plant-type reproduction factor. Curr Biol 18(8):607–613
Kelley LA et al (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10(6):845–858
Kumar S et al (2021) Plasmodium falciparum calcium-dependent protein kinase 4 is critical for male gametogenesis and transmission to the mosquito vector. MBio 12(6):e0257521
Kumar S et al (2022) A putative plasmodium RNA-binding protein plays a critical role in female gamete fertility and parasite transmission to the mosquito vector. Front Cell Dev Biol 10:825247
Sapir A et al (2007) AFF-1, a FOS-1-regulated fusogen, mediates fusion of the anchor cell in C. elegans. Dev Cell 12(5):683–698
Avinoam O et al (2011) Conserved eukaryotic fusogens can fuse viral envelopes to cells. Science 332(6029):589–592
Gattegno T et al (2007) Genetic control of fusion pore expansion in the epidermis of Caenorhabditis elegans. Mol Biol Cell 18(4):1153–1166
Mohler WA et al (2002) The type I membrane protein EFF-1 is essential for developmental cell fusion. Dev Cell 2(3):355–362
Podbilewicz B et al (2006) The C. elegans developmental fusogen EFF-1 mediates homotypic fusion in heterologous cells and in vivo. Dev Cell 11(4):471–481
Shemer G et al (2004) EFF-1 is sufficient to initiate and execute tissue-specific cell fusion in C. elegans. Curr Biol 14(17):1587–1591
von Besser K et al (2006) Arabidopsis HAP2 (GCS1) is a sperm-specific gene required for pollen tube guidance and fertilization. Development 133(23):4761–4769
Fedry J et al (2018) Evolutionary diversification of the HAP2 membrane insertion motifs to drive gamete fusion across eukaryotes. PLoS Biol 16(8):e2006357
Hernandez JM, Podbilewicz B (2017) The hallmarks of cell-cell fusion. Development 144(24):4481–4495
Liu Y et al (2015) The cytoplasmic domain of the gamete membrane fusion protein HAP2 targets the protein to the fusion site in Chlamydomonas and regulates the fusion reaction. Development 142(5):962–971
Okamoto M et al (2016) Two HAP2-GCS1 homologs responsible for gamete interactions in the cellular slime mold with multiple mating types: Implication for common mechanisms of sexual reproduction shared by plants and protozoa and for male-female differentiation. Dev Biol 415(1):6–13
Balmer EA, Faso C (2021) The road less traveled? Unconventional protein secretion at parasite-host interfaces. Front Cell Dev Biol 9:662711
Moi D et al (2022) Discovery of archaeal fusexins homologous to eukaryotic HAP2/GCS1 gamete fusion proteins. Nat Commun 13(1):3880
Vaughan JA (2007) Population dynamics of Plasmodium sporogony. Trends Parasitol 23(2):63–70
Sauerwein RW, Bousema T (2015) Transmission blocking malaria vaccines: assays and candidates in clinical development. Vaccine 33(52):7476–7482
Vermeulen AN et al (1985) Sequential expression of antigens on sexual stages of Plasmodium falciparum accessible to transmission-blocking antibodies in the mosquito. J Exp Med 162(5):1460–1476
Blagborough AM, Sinden RE (2009) Plasmodium berghei HAP2 induces strong malaria transmission-blocking immunity in vivo and in vitro. Vaccine 27(38):5187–5194
Miura K et al (2013) Functional comparison of Plasmodium falciparum transmission-blocking vaccine candidates by the standard membrane-feeding assay. Infect Immun 81(12):4377–4382
Angrisano F et al (2017) Targeting the conserved fusion loop of HAP2 inhibits the transmission of Plasmodium berghei and falciparum. Cell Rep 21(10):2868–2878
Feng J et al (2021) Structural basis of malaria transmission blockade by a monoclonal antibody to gamete fusogen HAP2. Elife 10:e74707
Trager W, Jensen JB (1976) Human malaria parasites in continuous culture. Science 193(4254):673–675
Carter R, Beach RF (1977) Gametogenesis in culture by gametocytes of Plasmodium falciparum. Nature 270(5634):240–241
Ifediba T, Vanderberg JP (1981) Complete in vitro maturation of Plasmodium falciparum gametocytes. Nature 294(5839):364–366
Zhang C et al (2014) Efficient editing of malaria parasite genome using the CRISPR/Cas9 system. MBio 5(4):e01414-e1514
Goswami D et al (2020) A replication-competent late liver stage-attenuated human malaria parasite. JCI Insight 5(13):e135589
Fidock DA, Wellems TE (1997) Transformation with human dihydrofolate reductase renders malaria parasites insensitive to WR99210 but does not affect the intrinsic activity of proguanil. Proc Natl Acad Sci USA 94(20):10931–10936
Azuma H et al (2007) Robust expansion of human hepatocytes in Fah−/−/Rag2−/−/Il2rg−/− mice. Nat Biotechnol 25(8):903–910
Vaughan AM et al (2015) Plasmodium falciparum genetic crosses in a humanized mouse model. Nat Methods 12(7):631–633
Dunsing V et al (2018) Optimal fluorescent protein tags for quantifying protein oligomerization in living cells. Sci Rep 8(1):10634
Funding
This work was funded by the NIH P01 AI127338 to A.M.V. and S.H.I.K., Seattle Children’s internal seed funding to S.H.I.K and the Israeli Science Foundation F.I.R.S.T No. 2327/19 to B.P. Funders had no role in study design, data collection, and interpretation or the decision to submit the work for publication.
Author information
Authors and Affiliations
Contributions
SK, SHIK and BP planned and analyzed experiments and wrote the manuscript. SK, CV, MTH, XL, KF, NGB, SYK; BAA, ASL, and NMC performed experiments. AMV, SHIK and BP acquired funding. SK, CV, NGB, BP, and SHIK analyzed the data. SK, CV, NGB, and MTH contributed to preparation of figures. The final manuscript was edited and approved by all authors.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing financial and non-financial interests.
Ethical statement
The animal experiments, which involved antisera generation in rabbits, were performed at Antagene Inc. (CA, US) and prescribed guidelines were followed.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file1 (MOV 5862 KB)
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kumar, S., Valansi, C., Haile, M.T. et al. Malaria parasites utilize two essential plasma membrane fusogens for gamete fertilization. Cell. Mol. Life Sci. 79, 549 (2022). https://doi.org/10.1007/s00018-022-04583-w
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-022-04583-w