Abstract
N6-methyladenosine (m6A) regulates many aspects of RNA metabolism and is involved in learning and memory processes. Yet, the impact of a dysregulation of post-transcriptional m6A editing on synaptic impairments in neurodegenerative disorders remains unknown. Here we investigated the m6A methylation pattern in the hippocampus of Huntington’s disease (HD) mice and the potential role of the m6A RNA modification in HD cognitive symptomatology. m6A modifications were evaluated in HD mice subjected to a hippocampal cognitive training task through m6A immunoprecipitation sequencing (MeRIP-seq) and the relative levels of m6A-modifying proteins (FTO and METTL14) by subcellular fractionation and Western blot analysis. Stereotaxic CA1 hippocampal delivery of AAV-shFTO was performed to investigate the effect of RNA m6A dysregulation in HD memory deficits. Our results reveal a m6A hypermethylation in relevant HD and synaptic related genes in the hippocampal transcriptome of Hdh+/Q111 mice. Conversely, m6A is aberrantly regulated in an experience-dependent manner in the HD hippocampus leading to demethylation of important components of synapse organization. Notably, the levels of RNA demethylase (FTO) and methyltransferase (METTL14) were modulated after training in the hippocampus of WT mice but not in Hdh+/Q111 mice. Finally, inhibition of FTO expression in the hippocampal CA1 region restored memory disturbances in symptomatic Hdh+/Q111 mice. Altogether, our results suggest that a differential RNA methylation landscape contributes to HD cognitive symptoms and uncover a role of m6A as a novel hallmark of HD.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Huntington's disease (HD) is an autosomal dominant neurodegenerative disorder that typically develops in young-middle adulthood. This disease is characterized by choreiform movements that appear in the later, more advanced disease stages, and by cognitive deficits and psychiatric symptoms that often appear at early stages even before the onset of motor symptoms [1, 2]. Though these impairments are commonly attributed to corticostriatal dysfunction, accumulating evidence demonstrates that memory decline in HD is likely a reflection of a widespread brain circuitry defect that involves hippocampal dysfunction [3,4,5]. Accordingly, HD patients present alterations in associative learning, spatial short-term memory, spatial working memory and recognition memory, which all are known to involve the participation of the hippocampus and temporal lobe structure [3, 6, 7].
Studies in HD mouse models have revealed that these early cognitive deficits better associate with synaptic dysfunction rather than with neuronal cell loss [8,9,10,11,12,13]. To understand this neuronal dysfunction, these studies have focused on the control of gene expression at the transcriptional level by transcription factors or epigenetic modifications of DNA or histones [14,15,16]. However, post-transcriptional mechanisms such as the regulation of miRNA expression, mis-splicing and altered polyadenylation [17,18,19,20,21] have also been described to be critical in the regulation of gene expression in HD. Notably, all these processes can be regulated by RNA modifications, in particular N6-methyladenosine (m6A), the most abundant internal modification in mRNA that is especially enriched in the mammalian brain [22, 23].
The m6A mark can be dynamically incorporated in the RNA by a methyltransferase complex consisting of METTL3, METTL14 and WTAP [24, 25] and removed by the demethylases FTO and ALKBH5 [26,27,28]. Transcriptome-wide mapping of m6A revealed that this modification is mainly deposited at the DRACH (where D = A, G or U; H = A, C or U) consensus motif displaying a conserved pattern across mRNAs and lncRNAs [23, 29]. A range of m6A binding proteins have been reported to mediate multifaceted functions of mRNA metabolism including splicing, nuclear export, stability, translation, microRNA processing, and subcellular targeting of m6A-modified mRNAs [30,31,32,33,34]. In the mammalian brain, m6A is also important in the fine-tuning of the transcriptome during learning and memory consolidation, particularly in the hippocampus, striatum and prefrontal cortex [35,36,37,38,39]. Indeed, m6A has been found to be present in the synaptic transcriptome, selectively modifying a number of transcripts in all cellular domains of tripartite synapses [40]. The possibility that dysregulation of m6A modifications could contribute to the disruption of the mRNA metabolism in neuronal dysfunction has already been suggested; however, only few studies have addressed its importance in neurodegenerative disorders [41,42,43,44,45,46].
In the present study, we aimed to investigate the potential role of m6A RNA modifications in HD cognitive deficits. Our data indicate that m6A methylation changes occur in the RNA along HD progression and that dysregulated m6A RNA modifications are involved in HD cognitive disturbances. Taken together, our findings identify m6A perturbations as a novel mechanism involved in memory impairments in HD, proposing this RNA modification as a potential therapeutic target for HD treatment.
Methods
Animals
HdhQ7/Q7 wild-type mice and heterozygous Hdh+/Q111 knock-in mice [47, 48] were bred and maintained on a C57BL/6 genetic background. Hdh+/Q111 mice present a targeted insertion of 109 CAG repeats in the murine huntingtin gene that extends the resulting polyglutamine segment to 111 residues. At 6 months of age, heterozygous mice present a HD-like phenotype with hippocampal long-term memory deficits, followed by motor coordination impairments at 8 months of age [13, 47]. To obtain age matched WT and Hdh+/Q111 littermates, male WT mice were crossed with female heterozygous Hdh+/Q111 mice. Only males from each genotype were used for the experimental procedures.
Behavioral training
We trained 4–5- and 8-month-old WT and Hdh+/Q111 mice (pre-symptomatic and symptomatic disease stages, respectively) in the object location task (OLT) to induce hippocampal neuronal activity. Mice were first habituated to an open field in the absence of objects and spatial cues for two consecutive days (10 min/day). This open field consisted of a white open-top squared arena: 40 × 40 × 40 cm. The third day, mice were placed in the open field with two identical objects and four different black spatial cues on each wall. Mice were allowed to explore for 10 min and then returned to their home cage. 30 min after the training task, mice were sacrificed by cervical dislocation.
Stereotaxic surgery and viral transduction in vivo
To knock down FTO expression, mice were stereotaxically injected with AVVs encoding eGFP scramble control shRNA or shFTO generated by Vector Biolabs. After anaesthesia with a mixture of oxygen and isoflurane, bilateral injection of AVVs (0.5 µl: 3.8 × 109 GC/hemisphere) in the CA1 of the dorsal hippocampus was performed according to the following coordinates from the bregma (millimetres); antero-posterior − 2.0; Lateral ± 1.5, and dorsoventral: − 1.3 (CA1). Injection was carried out as previously described [13]. Animals were carefully monitored for 2 h and then returned to their home cage for 1 month. After this period, animals from the different experimental groups were subjected to behavioral assessment.
Behavioral assessment
Animals were tracked and recorded with SMART Junior software (Panlab, Spain). Percentage of preference was calculated as (time exploring relocated object or new object)/(total time exploring both objects) × 100.
Object location task
OLT was performed as previously described [13]. During the acquisition phase on the third day, mice were presented with two identical objects (A and A’) placed in two adjacent corners of the experimental apparatus for 10 min, after which they were returned to their home cage. Twenty-four hours after the training phase, one of the objects was moved to the opposite corner, and mice were tested for 10 min.
Novel object recognition test
NORT was performed 3 days after the OLT as previously described [13]. Mice were presented with two identical objects on the third day (B and B´). After a delay of 24 h, animals were tested in the arena with a familiar object and a new object (B and C) placed in the same location, which they were allowed to explore for 10 min.
RNA isolation
RNA from the hippocampus was extracted using the RNeasy® Lipid Tissue Mini Kit from Qiagen® (cat no. 74804), following the instructions of the manufacturer. Concentration of purified RNA eluted in nuclease-free H2O was measured using the Nanodrop 1000 spectrophotometer (Thermo Fisher Scientific®).
Global m6A measurements
Total hippocampal m6A levels were assessed using the EpiQuik® m6A RNA Methylation Quantification Kit (Epigentek®, cat no. P-9005) and LC–MS/MS. EpiQuik was performed according to the manufacturer’s recommendations. RNA mass spectrometry was performed as previously described [49]. Mass spectrometric detection was performed using an Agilent 6495 Triple Quadrupole system.
MeRIP-seq
m6A immunoprecipitation sequencing was performed following the refined protocol of Zeng et al. [50], with minor modifications. Briefly, 3 µg of total hippocampal RNA from 3 animals/condition were chemically fragmented to 150–200 nt. Ten percent of the fragmented RNA sample was set aside to serve as the input and the remainder was then used to proceed with the IP. For this purpose, magnetic Protein A Dynabeads® (Invitrogen® by Thermo Fisher Scientific®, cat no. 10002D) were tumbled for 6 h with 5 µg of rabbit polyclonal anti-m6A antibody (Millipore®, cat no. ABE572) in IP buffer at 4ºC. After incubation, the antibody–beads mixture was washed and incubated overnight with 500 µL of the IP reaction mixture containing 9 µL of the fragmented RNA, 100 µL of 5× IP buffer and 10µL of SUPERase-IN® RNase Inhibitor (Invitrogen® by Thermo Fisher Scientific, cat no. AM2696) with constant rotation at 4 ºC. The immunoprecipitated RNA was subjected to two rounds of competitive elution with an m6A containing buffer and the eluted RNA was then concentrated using the RNeasy® MinElute® Cleanup kit (Qiagen®, cat no. 74204). Library construction was performed following the instructions provided by the SMARTer® Stranded Total RNA-Seq kit v2- Pico Input Mammalian (Takara Bio®, cat no. 634412). Dual combinatorial indexes were assigned to each sample to allow for multiplexing. After normalization of the concentration, libraries were pooled volumetrically and sequenced on the NovaSeq platform [Illumina®, BGI (Hong Kong)] performing paired end sequencing (2 × 150 bp).
MeRIP-seq data analysis
Quality control and adapter trimming
Coverage of the sequenced libraries was ~ 100 million reads (Q20% > 97 and Q30% > 93). Of those, the percentage of uniquely mapped reads was in the range of 65–85%, with 5% of multi-mapped reads. Adapter trimming was performed using Bbduk tool (BBtools—http://jgi.doe.gov/data-and-tools/bb-tools/), and the adapter.fa file obtained from BBTools (containing Illumina adapters). Additional trimming parameters were ktrim = r (indicating trimming from the right side), with Kmer size of 23, mink = 11 and hdist = 1. The command tbo was given to trim the adapter if it was based on pair overlap, and further tpe command allowed to trim both reads of the paired end sequencing to the same length.
Alignment of reads
Alignment of all the paired end reads with the mouse genome assembly mm9 was performed using STAR [51]. Samtools [52] view command was used to convert bam files to sam files. Further, to pick only uniquely mapped reads, Samtools view -q 255 filter was applied. Removal of repetitive regions was performed by filtering out the reads using bedtools intersect command [53].
Calculation of coverage
To count the reads mapping with all the overlapping features, quantification of the genes was performed using bedtools multicov. To obtain aligned reads, bam files belonging to all the replicates of an experimental condition were merged with the help of samtools merge command and reproducible regions were obtained from merged replicates.
Detection of m6A sites
Narrow peaks of each MeRIP/input sample pair were called using MACS2 [54] callpeak. We considered peaks that are present in at least two of the three replicates as reproducible, hence two out of three replicates were combined, using bedtools and irreproducible discovery rate (IDR). Further, IDR [55] was used to check the reproducibility between the replicates taking p-value as a ranking measure. Combined peaks were filtered to find significant peak calls using IDR score filter (< = 0.05). Coordinates of all of the reproducible peaks in combination of two were concatenated and later collapsed using mergeBed [53]. Annotation of the reproducible merged peaks with the gencode.vM1.annotation.gtf file was performed using intersectBed [53].
Differential peak analysis, annotation and enrichment analysis
Differential peak analysis was performed between conditions with the reproducible peaks, using a DiffBind tool (annotationdbi) [56]. Statistically differential peak-sets were obtained using DESeq2 and filtered using p value < 0.05. Adjusted p-value was obtained by Benjamin and Hochberg multiple testing adjustment. Further, annotation of the differential peaks to the corresponding genes was performed with bedtools intersect using mm9.knownGene.bed. Annotation of gene biotypes (3′/5′ UTR, CDS exons and introns) of differentially bound peaks was performed using ChIPseeker [57].
Gene ontology
For gene ontology (GO), significant differential peaks were analyzed with Ingenuity Pathway Analysis (IPA) software (QIAGEN—http://www.ingenuity.com) and Synaptic Gene Ontologies (SynGo) [58].
Motif identification
Motif analysis was run de novo using HOMER (version 4.11) [59], considering ± 100 nucleotide intervals around the peak summit of ~ 3500 best scoring reproducible peaks in the 3’UTR. Background sequences were generated from respective input sequences using the scrambleFasta.pl script. Next, de novo motif search was run using findMotifs.pl script with -rna -p 10 -len 6 as parameters.
Differential gene expression
For analysis of RNA expression, readcounts from input samples were used applying a CPM cutoff of 1 or above in all three biological replicates [60] to discriminate expressed genes for the entire dataset. Genes were normalized using TMM algorithm [60] and calcNormFactor of edgeR [61]. The relationship function voom [62] from the limma package was used to establish the mean variance relationship and generate weights for each observation. The lmFit function of limma was used to transform the RNA-Seq data before linear modeling and find differentially expressed (DE) genes.
Subcellular fractionation and western blot analysis
Nuclear enrichment was performed as described previously [13]. Enrichment of the nuclear fraction was assessed by the presence of the specific nuclear marker Histone H3.
Synaptosomal fractionation was performed as described elsewhere in Bruyere et al. [63]. Enrichment of each fraction was assessed by the presence or absence of specific synaptosomal markers: synaptophysin (non-PSD fraction) and postsynaptic density protein 95 (PSD95) (PSD fraction).
A standard Western blot protocol was employed to quantify protein levels from the subcellular fractionations with the following antibodies: FTO (1:1000, NovusBio, cat nº NB110-60,935), METTL14 (1:2000, Sigma-Aldrich, cat nº HPA038002), PSD-95 (1:1000, Cell Signalling Technology, cat no. 3450), synaptophysin (1:1000, Synaptic Systems, cat no. 101011), Histone H3 (1:1000, Cell Signaling Technology). Immunoreactive bands were developed by the enhanced chemiluminiscence method, detected using ChemiDoc imaging system (Bio-Rad®) or films. ImageLab® Software Version 6.0 (2017) or Image J were used for quantification.
Tissue fixation, immunofluorescence and confocal imaging
Animals were deeply anaesthetized and immediately perfused intracardially with 4% (weight/vol) paraformaldehyde in 0.1 M phosphate buffer. Immunofluorescence was performed as previously described [13]. Following permeabilization and blocking, free floating brain sections were incubated overnight at 4 ºC with the primary antibodies against FTO (1:200, NovusBio, cat no. NB110-60935) and m6A (1:200, Synaptic Systems, cat no. 202 111). Samples were incubated with appropriate secondary antibodies (1:200, Cy3-coupled fluorescent secondary antibody, Jackson ImmunoResearch Laboratories, cat no. 715-165-150; 1:200, Alexa Fluor 647 goat anti-mouse, Invitrogen, cat no. A21236) for 2 h at room temperature. Nuclei were stained for 10 min with 4′,6-diamidino-2-phenylindole (DAPI) (1:5000, Sigma Aldrich, cat nº D9542). Representative images from the CA1 nuclear layer were obtained with a Leica SP5 laser scanning confocal microscope (Leica). Integrated optical density of nuclear FTO was analyzed with Image J.
Statistical analysis
Raw data were processed using Excel® Microsoft Office and for further analysis transferred to Graphpad Prism® version 8.0.2 for Windows. Results are expressed as mean ± SEM. Normal distribution was assessed with the Shapiro–Wilk test. For statistical analysis, unpaired Student’s t-test (two-tailed) or two-way ANOVA was performed, and the appropriate post-hoc tests were applied as indicated in the figure legends. A 95% confidence interval was used, considering differences statistically significant when p < 0.05.
Results
Landscape of m6A in the Hdh +/Q111 mouse hippocampus under naive and cognitive-trained conditions
Previous studies have shown that dynamic regulation of m6A on mRNA confers transcriptome plasticity while altered m6A methylation severely impacts gene expression and impairs learning and memory [35,36,37,38,39,40]. Therefore, we sought to evaluate whether HD mutation alters the m6A profile on mRNA under naive and cognitive training conditions in Hdh+/Q111 heterozygous mice. Behavioral analysis of mice subjected to the training session in the spatial task (Fig. 1a) showed no significant differences between genotypes at 5 months of age while at 8 months of age Hdh+/Q111 mice reproduced the well-described defects in cognition and motor coordination (Supplementary Fig. 1a–f) [13, 47]. Next, global m6A levels were assessed in total hippocampal RNA from naive mice by liquid chromatography-tandem mass spectrometry (LC–MS/MS). We found increasing m6A levels in hippocampal RNA from Hdh+/Q111 mice when compared to WT mice (Supplementary Fig. 2a). Similar upregulation under naive conditions was observed when m6A levels were further measured using the EpiQuik m6A RNA Methylation Quantification Kit. Interestingly, we observed that following training, similar m6A levels were observed in WT mice compared to naive conditions while a significant decrease was found in Hdh+/Q111 mice (Supplementary Fig. 2b).
Mapping of the transcriptome-wide m6A landscape in the hippocampus of WT and Hdh+/Q111 (KI) mice. A Schematic illustration of the experimental design used to induce hippocampal activity. Mice were habituated to the open-field arena for two consecutive days. At the third day mice were trained on the OLT for 10 min and sacrificed after 30 min for hippocampal dissection to perform global m6A measurement, MeRIP-seq and Western Blot analysis. Metagene plot showing the m6A peak distribution along the mRNA in 5- (B) and 8- (C) month-old mice. Peaks are enriched at the CDS and 3′UTR in all conditions. Sequence logo representing the deduced consensus motif associated with m6A peaks identified by HOMER in 5- (D) and 8- (E) month-old mice. Histograms depict the number of reproducible m6A peaks in each condition (left) and the number of m6A modified RNAs (right) in 5- (F) and 8- (G) month-old mice. IDR irreproducible discovery rate, KI knock-in
To further identify and characterize the m6A RNA modification pattern, m6A RNA immunoprecipitation and sequencing (MeRIP-seq) was performed. The distribution of the peaks is represented as a metagene plot in Fig. 1b and c, showing a similar enrichment of m6A density at the coding sequence (CDS) and 3′-untranslated regions (UTR) in all conditions. Analysis of the sequence context around called peaks showed enrichment of the m6A consensus site DRACH in the different conditions analyzed (Fig. 1d and e). These data indicate that our m6A profiling results are in accordance with the published m6A features [23, 29]. MeRIP-seq analysis identified around 30,000 m6A peaks for each of the conditions corresponding to around 11,000 m6A modified mRNAs (Fig. 1f and g; Supplementary table 1). Consistent with our findings showing increased m6A levels in RNA from naive symptomatic Hdh+/Q111 mice (Supplementary Fig. 2), the number of m6A peaks in these mice was 1.4-fold higher compared to WT mice (Fig. 1f and g).
Altered transcriptome methylation in Hdh +/Q111 mouse hippocampus affects synaptic and HD-related genes at pre- and symptomatic disease stages
In order to characterize basal m6A methylation signatures along the disease progression, we analyzed the differential m6A methylation peaks between naive WT and Hdh+/Q111 mice by a differential enrichment analysis at 5 and 8 months of age (Supplementary Tables 2 and 3). Adjusted p value < 0.05 and log2fold change > 1 or < − 1 were used as screening threshold of differential peaks. At 5 months of age, 270 peaks were hypermethylated and 149 peaks were hypomethylated in Hdh+/Q111 mice when compared to WT mice, while at 8 months of age 417 peaks were hypermethylated and 109 hypomethylated (Fig. 2a and b). When comparing all the differential peaks observed at pre-symptomatic and symptomatic stages, we identified only 5% of overlapping m6A tagged genes, while 42% and 53% of the differential methylated genes were unique for each stage (Fig. 2c), indicating a unique pattern of pathological methylation at each disease stage. To investigate the association between m6A methylation and mRNA expression, we intersected differential mRNA expression with differential m6A peaks. At 5 months of age, we found a greater number of m6A-containing mRNAs with significant differences in mRNA expression (47%) than at 8 months of age (23%) (Supplementary Fig. 3a), although no significant correlation between down and upregulation of mRNA species and m6A enrichment was found (Supplementary Fig. 3b and c). At both stages, we identified changes in the levels of m6A in different genes linked to HD, such as Pde10a and Eif3b (at 5 months), Kalrn, Ntrk2, Gnaq, Grin2b, Dyrk1a (at 8 months) and Htt itself (at 5 and 8 months), thus strengthening our hypothesis that altered m6A methylation plays a role in HD pathology (Tables 1 and 2). We also detected significant changes in m6A methylation in synaptic genes such as Nefl (at 5 months) and Slc6a17, Frx1 (at 8 months) (some representative genes are shown in Fig. 2d).
Mutant huntingtin induces alterations in hippocampal m6A RNA modifications affecting synaptic and HD-related genes. Volcano plots of the differentially methylated peaks between naive KI and WT animals at 5- (A) and 8- (B) months of age. Names of representative genes are indicated. C Venn diagram of the overlap of differential m6A modified transcripts between naive KI and WT mice at 5- and 8- months of age. Only 5% of differential m6A marked transcripts overlap between pre- and symptomatic disease stages. D UCSC Genome browser tracks of representative differential m6A peaks along indicated mRNAs. The y-axis represents normalized number of reads. Blue thick boxes represent the open reading frame while the blue line represents the untranslated regions (arrows point to the direction of transcription). Grey reads are from WT animals, black reads are from KI animals. Scale bar 1 kb. Distribution analysis of the down- and up-regulated differential m6A peaks between naive KI and WT mice at 5- (E) and 8- (F) months of age along the four transcript segments: 5′UTR, CDS exon, 3’UTR, intron. Data are shown as percentage of total differential peaks. IPA of differentially hypermethylated transcripts between naive KI and WT mice at 5- (G) and 8- (H) months of age. Bar plots showing the -Log (Adj. p value) of enriched terms belonging to the different categories. Screening threshold: Adjusted p value < 0.05 and log2 fold change > 1 or < − 1. FC fold change
Since the unique distribution of m6A along the transcripts provides hints as to its functions [23, 29], we determined the location of the up and downregulated m6A peaks in Hdh+/Q111 mice at 5 and 8 months of age. We divided the transcript into four areas: 5'UTR, 3′UTR, CDS exon, and intron (Fig. 2e and f). All the differential m6A peaks showed similar distribution at 5 and 8 months, with a slight but not significant increase of upregulated peaks in the 3′ UTR and a decrease of upregulated peaks in the 5´UTR at 8 months compared to 5-month-old mice (Supplementary Fig. 3d). Moreover, we found around 40% of the m6A peaks located in introns. Since m6A changes may also influence RNA splicing and polyadenylation [64,65,66,67], we next explored whether the observed alterations in methylation in Hdh+/Q111 mice could be related with known genes affected by mis-splicing or aberrant polyadenylation in HD [18, 21]. We integrated our dataset with published datasets of alternative splicing and polyadenylation analysis from the R6/1 HD mouse model at symptomatic disease stages (3,5 and 7–8 months, respectively) [18, 21]. Integration analysis showed a significant over-enrichment of genes with changes in m6A levels and genes described to be mis-spliced, both at 5 and 8 months of age (Supplementary Fig. 4a, b and Supplementary Tables 4 and 5). Notably, the majority of the m6A peaks present in overlapping methylated and miss-spliced genes were found in introns and exon (Data not shown). At 8 months, we also detected a significant overlap between genes with m6A changes and genes described to have an altered poly(A) tail length in 7–8 month-old R6/1 mice (Supplementary Fig. 4c and d and Supplementary Tables 6 and 7). These results suggest that alteration of m6A methylation in hippocampal genes could be associated to aberrant alternative splicing and/ or polyadenylation in HD.
To analyze whether these changes in the m6A landscape along disease progression are associated to distinct functional signatures, we performed Ingenuity Pathway Analysis (IPA) with the lists of all transcripts differentially hypo- or hypermethylated between Hdh+/Q111 and WT mice at 5 and 8 months of age. Top diseases and bio functions obtained with IPA are given in Supplementary Fig. 5a–d. IPA for diseases and disorders revealed an enrichment in neurological diseases specifically in the hypermethylated transcripts, both at 5 and 8 months of age. Likewise, among molecular and cellular functions, terms related to cells morphology including “microtubule dynamics” and “formation of cellular protrusions,” were particularly enriched in hypermethylated transcripts at 5 months of age, while “synaptic depression,” “neurotransmission,” “formation of cellular protrusions” as well as “processing of mRNA,” among others, were enriched in hypermethylated transcripts at 8 months (Fig. 2g, h and Supplementary Tables 8 and 9 for complete lists of all biological functions and their associated genes). The top physiological system development and functions at 8 months of age were “cognition,” “learning” and “locomotion.” These processes have been described to be influenced by mutant huntingtin toxicity [2, 68,69,70,71,72] and our findings suggest that this is in part mediated by altered regulation of m6A post-transcriptional mRNA modifications.
Our data revealed differential m6A methylation patterns between WT and Hdh+/Q111 mice along the disease progression. Next, we sought to investigate whether mutant huntingtin expression was associated with changes in methylation dynamics in hippocampal cognition. To this aim, we analyzed the differential methylated peaks between Hdh+/Q111 and WT mice after the spatial training in the OLT. We found 277 hypermethylated m6A peaks and 170 hypomethylated m6A peaks at 5 months of age in trained Hdh+/Q111 mice compared to WT mice, while at 8 months only 165 peaks were hypermethylated whereas 392 peaks were hypomethylated (Fig. 3a, b and Supplementary Tables 10 and 11). To deepen about the functional consequences of these changes, IPA pathway analysis between trained Hdh+/Q111 and WT mice was performed (Supplementary Tables 12 and 13 for complete lists of all biological functions and their associated genes), analyzing hypo- and hypermethylated transcripts independently. We found that the strongest enrichment in relevant functional terms was obtained in the hypomethylated transcripts, with an important enrichment in “motor or movement disorder,” including “Huntington´s disease” within both ages (Fig. 3c, d and Supplementary Fig. 6a–d). Importantly, at 8 months of age, training in Hdh+/Q111 mice induced the hypomethylation of genes tightly involved in synaptic transmission when compared to WT trained mice (Fig. 3d). No significant terms could be detected in the hypermethylated transcripts at 8 months, and only a few interesting terms were found in hypermethylated transcripts at 5 months (Data shown in supplementary tables 12 and 13).
Behavioral training induces differential methylation of synaptic genes in symptomatic Hdh+/Q111 (KI) mice. Volcano plots of the differentially methylated peaks between trained KI and WT animals at 5- (A) and 8- (B) months of age. Names of representative genes are indicated. IPA of hypomethylated transcripts between trained KI and WT mice at 5- (C) and 8- (D) months of age. Bar plots showing the -Log (adj. p value) of enriched terms belonging to the different categories. Screening threshold: adjusted p value < 0.05 and log2 fold change > 1 or < − 1
Symptomatic Hdh +/Q111 mice show aberrant m6A demethylation after hippocampal cognitive training.
Our previous data have revealed altered m6A RNA patterns between WT and Hdh+/Q111 mice at pre- and symptomatic stages, both in basal and after cognitive engagement conditions. However, when comparing Hdh+/Q111- vs WT-trained mice, we observed a stronger enrichment of differentially methylated genes on the “Synaptic Transmission” function in symptomatic disease stages. To get further insight into the specific changes in m6A associated with cognitive training, we compared the differential m6A peaks in training vs naive conditions within each genotype at 8 months of age (Supplementary Tables 14 and 15). Using the same filtering criteria as above (Fig. 2a, b) we identified in trained vs naive WT mice, 473 hypermethylated m6A peaks and 221 hypomethylated m6A peaks (Fig. 4a). In contrast, when trained Hdh+/Q111 mice were compared to naive Hdh+/Q111 mice, the differential analysis revealed only 168 hypermethylated m6A peaks and 359 hypomethylated m6A peaks (Fig. 4b). The extent of overlap among differential methylation in each genotype at 8 months of age in response to training is shown in Fig. 4c, d. We observed a unique pattern of hypermethylation and hypomethylation in each genotype, with only a minority of differential m6A marked transcripts in common between WT and Hdh+/Q111 mice. In addition, the extent of m6A changes of differential m6A-marked transcripts identified in WT did not resemble the methylation pattern observed in KI (Fig. 4e), re-enforcing the idea that memory-induced m6A methylation is impaired in KI mice. Moreover, the hierarchical heatmap representation of differential methylated transcripts in common (Supplementary Fig. 7a, b) showed a cluster of Immediate-Early Genes (IEGs) which are similarly hypermethylated after the training task (Fig. 4b), in line with previous evidence showing increased methylation of IEGs after behavioral training [39] and a cluster of 34 transcripts with opposite genotype-dependent m6A regulation.
Symptomatic Hdh+ /Q111 (KI) mice exhibit prominent demethylation in response to behavioral training. Volcano plots of the differentially methylated peaks between trained and naive conditions in WT (A) and KI (B) 8-month-old animals. Names of representative genes are indicated. Venn diagram showing shared and unique differential (C) hypermethylated and (D) hypomethylated transcripts in WT and KI after training. E Heatmap representing the fold change of m6A methylation levels between trained and naive conditions in differentially methylated transcripts identified in WT and KI animals. Distribution analysis of the down- and up-regulated differential m6A peaks between naive and trained conditions in symptomatic disease stages in WT (F) and KI (G) mice along the four transcript segments: 5′UTR, CDS exon, 3′UTR, intron. Data are shown as percentage of total differential peaks. H UCSC Genome browser tracks showing representative differential m6A peaks (The y-axis represents normalized number of reads. Blue thick boxes represent the open reading frame while the blue line represents the untranslated regions (arrows point to the direction of transcription). Screening threshold: adjusted p value < 0.05 and log2 fold change > 1 or < − 1
When comparing trained versus naive conditions in both WT and Hdh+/Q111 mice, the percentage of m6A marked transcripts with differential mRNA expression was around 30% (Supplementary Fig. 8a), indicating that m6A might not affect RNA decay or stability in the remaining transcripts but could affect other aspects of RNA function or metabolism. The association between m6A methylation and mRNA expression in the m6A marked transcripts showed a positive and significant correlation between the magnitude of change in expression and the magnitude of m6A peak enrichment in upregulated mRNAs for both genotypes. In contrast, a statistically significant but weak correlation was found between the downregulated mRNA species and m6A peak enrichment, only for Hdh+/Q111 mice (Supplementary Fig. 8b and c). These results suggest that only the addition of m6A to RNA species in response to cognitive engagement correlates with their increased mRNA expression levels in both genotypes. We further analyzed the distribution of the differential m6A peaks along the transcripts. Here, Hdh+/Q111 and WT mice show an equal distribution in CDS exon, 5’UTR and 3′UTR, with the most frequently occurring position being within introns (Fig. 4f and g). Nevertheless, in trained WT mice we observed a higher percentage of upregulated m6A peaks in introns compared with trained Hdh+/Q111 mice, which in turn showed a higher percentage of downregulated m6A peaks (Supplementary Fig. 8d). To illustrate these results, four representative genes showing the m6A methylation pattern have been depicted (Fig. 4h).
Given the role of m6A methylation in synaptic signaling and potentiation during neuronal activation, we next explored the functional signature displayed by the differentially methylated transcripts using the Synaptic Gene Ontologies (SynGO) database. We observed that training-induced hypermethylated genes in WT mice were significantly enriched in “synaptic organization,” “synaptic signaling” and “presynaptic and postsynaptic processes” (Fig. 5a), when compared to enriched hypermethylated genes in Hdh+/Q111 mice (Fig. 5c). By contrast, in Hdh+/Q111 mice, synaptic processes involving “organization” and “synaptic signaling” were enriched for hypomethylated genes (Fig. 5b, d). A similar gene ontology signature of hypomethylation after behavioral training was observed in Hdh+/Q111mice at the age of 5 months, further supporting the idea that m6A methylation is altered during neuronal activity in Hdh+/Q111 mice (Supplementary Fig. 9a–f and Supplementary Tables 16 and 17). These results indicate that the HD mutation impacts on the regulation of m6A methylation, decreasing in synaptic mRNAs after cognitive engagement, which might critically contribute to synaptic and memory dysfunction in Hdh+/Q111 mice.
Functional analyses of synaptic genes and gene enrichment of differentially methylated transcripts in trained symptomatic Hdh+/Q111 (KI) mice. Sunburst plots for synaptic GO of differentially hypermethylated (A, C) and hypomethylated (B, D) genes in response to the OLT training in 8-month-old WT mice (A, B) and KI mice (C, D). In response to the behavioral training task, WT mice show an increased number of hypermethylated genes enriched in synaptic terms, whereas in KI mice genes enriched in synaptic terms are hypomethylated. Numbers in the center indicate number of genes enriched in synaptic terms. Tables below indicate most relevant synaptic genes. GO analysis was performed with the SynGo knowledgebase. Screening threshold: adjusted p value < 0.05 and log2 fold change > 1 or < − 1. GO gene ontology
Finally, since the acquisition of spatial memories during the cognitive training task is known to be modulated by activity-dependent modifications of cornu ammonis 1 (CA1) synapses [73], we performed data integration of MeRIP-seq data from 8-month-old WT and Hdh+/Q111 mice after training with data from a cell type-specific CA1 hippocampal transcriptomic database [74]. This analysis showed that at 8 months, transcripts enriched in CA1 neurons, which present structural and functional synaptic deficits in HD [11,12,13, 70], were highly and significantly overlapping with training-induced hypermethylated transcripts in WT mice, while a lower enrichment was found in Hdh+/Q111 mice hippocampus (48 genes enriched in CA1 neurons in WT mice and 11genes in Hdh+/Q111 mice) (Fig. 6a). In addition, hypomethylated transcripts were also associated with CA1 neuronal-enriched transcripts in WT hippocampus while Hdh+/Q111 hypomethylated transcripts were enriched in both CA1 neuronal and glial enriched transcripts after training (Fig. 6b). Together, these results indicate that m6A dysregulation affects CA1 neuronal transcripts in Hdh+/Q111 mice suggesting a role of m6A in transcriptional remodeling underlying acquisition of spatial memory.
Differential methylated transcripts in Hdh+/Q111(KI) mice in response to training are enriched in CA1 neurons. Multi-layer pie charts showing integration of hypermethylated transcripts (A) and hypomethylated transcripts (B) from 8-month-old WT and KI mice subjected to behavioral training with cell-type-specific CA1 hippocampal transcriptomic database. Outer levels indicate degree of correlation between the levels marked in green (indicated as − Log10 (p value)). The size of each item represents its contribution (number of m6A-modified transcripts) to the inner parent category. Screening threshold: adjusted p value < 0.05 and log2 fold change > 1 or < − 1
Subcellular expression of FTO is modulated in WT but not Hdh +/Q111 mice following a hippocampal cognitive training task
Regulation of m6A in response to sensory experience and learning is supported by methyltransferases and demethylases [28, 35, 75, 76]. To define whether altered m6A methylation patterns in Hdh+/Q111 mice were related with changes in the levels of m6A-modifying proteins, expression of METTL14 and FTO was analyzed in WT and Hdh+/Q111 mice hippocampus under naive and training conditions. Since both enzymes are enriched in the nucleus [25, 27], subcellular fractionation assays in hippocampal lysates were performed to determine the levels of nuclear METTL14 and FTO. At 5 months of age, similar levels of FTO and METTL14 were found in nuclear fractions between genotypes and conditions (Supplementary Fig. 10a–c). By contrast, at 8 months of age a significant increase of both proteins was observed in WT mice in response to hippocampal training while no significant changes were detected in Hdh+/Q111 mice (Fig. 7a–c). Since FTO and METTL14 have also been found in synapsis and distal neuronal processes [36, 40] and given the changes detected at 8 but not 5 months of age following training, we next analyzed the levels of both m6A regulatory enzymes in postsynaptic density (PSD) and non-PSD fractions (Fig. 7d–g). We observed that FTO levels decrease in the postsynaptic fraction in WT mice after training while no changes were observed in Hdh+/Q111 mice (Fig. 7d, e). Altogether, these findings indicate that at a symptomatic stage characterized by hippocampal cognitive deficits, Hdh+/Q111 mice display altered protein levels of FTO and METTL14.
Nuclear and post-synaptic levels of the m6A modifying proteins do not change upon training in symptomatic Hdh+/Q111 (KI) mice. Western Blots for METTL14 and FTO in A–C nuclear enriched fractions, D, E post-synaptic (PSD) and F, G non-PSD fractions from hippocampus of 8-month-old mice subjected to the training in the OLT. A–C Nuclear fraction was normalized to Histone H3. Two-way ANOVA reported a significant training effect in FTO levels (F(1,25) = 10.96, p = 0.0028) and in METTL14 levels (F(1,26) = 7.101, p = 0.0131) (n = 6–10 animals/condition). Right: Representative immunoblots D, E PSD fraction was normalized to PSD95. Two-way ANOVA reported a significant training effect in FTO levels (F(1,15) = 11.21, p = 0.0044) (n = 4–6 animals/condition). Below the graphs: Representative immunoblots (F, G). Non-PSD fraction was normalized to synaptophysin. (n = 5–7 animals/condition). Below the graphs: Representative immunoblots. Data were analyzed using Tukey as a post hoc test. *p < 0.05 and **p < 0.01 compared with WT naive mice. $p < 0.05 compared with WT-trained mice. Data are presented as mean ± SEM. N naïve, T trained, Syn synaptophysin
Inhibition of m6A demethylation in hippocampal CA1 region improves exploratory behavior and spatial memory in Hdh +/Q111 mice
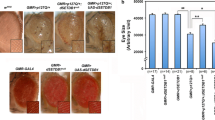
In view of the results showing decreased levels of m6A methylation in synaptic-related genes in trained Hdh+/Q111 mice, we hypothesized that synaptic and cognitive defects in HD mice could be related with a dysregulation of m6A modification dynamics. To test our hypothesis, we investigated the effect of FTO knockdown on the hippocampal-dependent memory deficits previously described in Hdh+/Q111 mice [13]. Adeno-associated viruses (AAV) encoding eGFP scramble control shRNA or shFTO were injected bilaterally into the dorsal hippocampus of WT and Hdh+/Q111 mice at 8 months (Fig. 8a). To test the efficiency of shRNA in knocking down FTO expression, we measured by immunofluorescence intensity the levels of FTO levels in the CA1 region following shFTO injection. A significant reduction of around 30% was found when compared to scramble control (Fig. 8b). Moreover, immunofluorescence co-labelling studies of FTO and m6A confirmed that m6A immunoreactivity was reduced in the pyramidal neurons of the CA1 in Hdh+/Q111 mice compared with WT mice while increased in Hdh+/Q111 mice following shFTO injection (Fig. 8c). To study the effect of FTO knockdown in the hippocampus, we first analyzed spatial memory by the OLT (Fig. 8d) [77, 78]. During training, all mice similarly explored both objects, indicating no object or place preferences between genotypes (Supplementary Fig. 11a and b). However, Hdh+/Q111 mice displayed decreased total exploration time (Supplementary Fig. 11c). When long-term spatial memory was assessed 24 h after training, Hdh+/Q111 mice injected with AAV control shRNA exhibited a significantly lower preference for the object displaced to the new location when compared to WT mice and Hdh+/Q111 mice injected with AAV shFTO (Fig. 8e), confirming previous results from our lab showing hippocampal-dependent deficits in Hdh+/Q111 mice [13, 16]. Importantly, FTO knockdown in Hdh+/Q111 mice lead not only to a significant preference for the new object location but also to an increase in total exploration time indicating that recovery of m6A levels in Hdh+/Q111 mice rescue spatial memory deficits (Fig. 8e, f). Similar results were obtained when we evaluated recognition memory by Novel Object Recognition Test (NORT) (Fig. 8g) [79]. No significant object preference between genotypes was found during the training period, while a significant decrease in total exploration time was observed in Hdh+/Q111 compared to WT mice (Supplementary Fig. 11d–f). Twenty-four hours after training, Hdh+/Q111 mice injected with AAV control shRNA exhibited a significantly lower preference for the novel object compared with WT mice and with Hdh+/Q111 mice injected with AAV shFTO (Fig. 8h), indicating preserved long-term recognition memory in Hdh+/Q111 mice with normalized levels of FTO and restored levels of m6A. As previously observed during OLT test, memory rescue was accompanied by an increase in exploratory behavior (Fig. 8i). Taken together, these data show that modulation of m6A levels in symptomatic Hdh+/Q111 mice by knockdown of FTO expression in the CA1 of the dorsal hippocampus restores HD spatial and recognition memory.
Targeted knockdown of FTO in the dorsal hippocampus of Hdh+/Q111 (KI) mice rescues spatial memory deficits at 8 months of age. A Representative image showing AAV injection site in the CA1. Immunological staining shows GFP (green) and DAPI (blue). Scale bar 150 µm. B Relative levels of nuclear FTO (Integrated Optical Density) in the CA1 of injected mice. Two-way ANOVA reported a significant treatment effect (shFTO) (F(1,128) = 92.73, p < 0.0001) (n = 3–4 animals/condition) (3 slices/animal were analyzed, 2 images for each slice). C Representative confocal images of dorsal CA1 transduced with sh-sc or shFTO in WT and KI mice. Triple staining showing FTO (red), m6A (yellow) and DAPI (blue). Scale bar 5 µm. Schematic illustration of OLT (D) and NORT (G) behavioral assessment. E Percentage of time exploring the displaced object for WT sh-sc, WT shFTO, KI sh-sc and KI shFTO. Two-way ANOVA reported a significant interaction effect (F(1,35) = 21.00, p < 0.0001), a significant treatment effect (shRNA) (F(1,35) = 5.819, p = 0.0212) and a significant genotype effect (F(1, 35) = 22.55, p < 0.0001). F Exploration time (sec) during the testing session of OLT. Two-way ANOVA reported a significant interaction effect (F(1, 36) = 11.63, p = 0.0016) and a significant treatment effect (shRNA) (F(1, 36) = 10.09, p = 0.0031. H Percentage of time exploring the new object for WT sh-sc, WT shFTO, KI sh-sc and KI shFTO. Two-way ANOVA reported a significant interaction effect (F(1,37) = 14.08, p = 0.0006), a significant treatment effect (shRNA) (F(1, 37) = 6.154, p = 0.0178) and a significant genotype effect (F(1,37) = 12.22, p = 0.0012). I Exploration time (sec) during the testing session of NORT for WT sh-sc, WT shFTO, KI sh-sc and KI shFTO. Two-way ANOVA reported a significant interaction effect (F(1,37) = 8.840, p = 0.0052). Dashed line shows chance levels for exploration. Two-way ANOVA with Tukey’s multiple comparisons test; *p < 0.05 and ***p < 0.001 compared with WT sh-sc mice; $p < 0.05 and $$$p < 0.001 compared with KI sh-sc mice. Data are presented as mean ± SEM (n = 9–12 per genotype). DG dentate gyrus
Discussion
Our study provides for the first time, evidence of m6A hypermethylation in relevant HD and synaptic-related genes in the hippocampal transcriptome of Hdh+/Q111 mice. We also found that m6A is aberrantly regulated in an experience-dependent manner in the HD hippocampus leading to demethylation of important components of synapse organization which could underlie HD cognitive deficits. In agreement with these data, we show that FTO knockdown in HD mice improves hippocampal spatial and recognition memories. Overall, these findings suggest that m6A RNA modifications represent a new RNA post-transcriptional signature underlying gene expression dysregulation and cognitive deficits in HD.
Here, we identified in a HD mice model, hypermethylation in several transcripts of genes previously described to be altered in HD such as, Pde10a [80, 81], Eif3b [82], Kalrn [47], Ntrk2 [83], Grin2b [84, 85], Dyrk1a [86] and Htt itself. These findings suggest a possible contribution of m6A modifications in the HD physiopathology. To date studies in Alzheimer´s and Parkinson´s Disease have suggested that m6A dysregulation is a common feature of many neurodegenerative diseases [42, 46] and have emphasized the importance of an appropriate m6A equilibrium for an accurate hippocampal function. Accordingly, we have shown that hypermethylated transcripts in symptomatic Hdh+/Q111 mice under naive conditions are enriched in synapse-related functions, supporting a role of m6A in mediating HD synaptic and memory deficits. This finding reveals a new layer of dysregulation in the expression of synaptic genes, a process previously associated in HD to changes in the activity or levels of transcription factors and epigenetic modifications [14,15,16, 87, 88]. Indeed, recent studies have revealed that m6A is present in the synaptic transcriptome, modifying in a selective manner different transcripts and shaping the synaptic proteome [40], which support our hypothesis that disruption or dysregulation of m6A modifications would impact memory processes in HD mice. In this regard, behavioral studies have demonstrated the activity-dependent nature of neuronal m6A in response to behavioral experience and memory formation in different brain regions [35, 36, 76]. In accordance, we found that mRNA methylation was actively regulated in the dorsal hippocampus of both WT and Hdh+/Q111 mice in response to spatial learning, though a different m6A pattern was observed in the transcriptome of Hdh+/Q111 mice when compared to WT mice. Thus, although both genotypes showed similar methylation in IEGs and synaptic genes known to be target of m6A modifications [39, 76, 89, 90], Hdh+/Q111 mice failed to increase methylation or induced demethylation of some synaptic genes critical for proper learning. Since an increase of m6A in the mouse hippocampus or prefrontal cortex in response to learning has been associated with the constrain of the sorting efficiencies or turnover of nascent RNAs, the reduced m6A methylation observed in Hdh+/Q111 hippocampus could indicate that the degradation and stability of several synaptic genes is affected in these mice.
An interesting finding of this study is the consistent and differentially methylated peak in intron 1 of mHtt, in both 5- and 8-month-old Hdh+/Q111 naive and trained mice. Remarkably, studies have shown that the increased number of CAG repeats in mHtt leads to a slower transcription rate of RNA polymerase II in this intronic region [91]. In turn, slow transcription speeds have been associated to an elevated m6A content [92] pointing to a possible link between the number of CAG repeats and m6A deposition in Htt intron. This evidence further strengthens the contribution of m6A in HD physiopathology, although its function on Htt RNA fate remains to be investigated.
RNA m6A methylation can have mixed effects beyond regulation of transcript abundance. Indeed, the specific location of m6A can determine alternative polyadenylation usage [65, 93], deadenylation-mediated RNA degradation [64], and alternative splicing [34, 67]. In this sense, when comparing naive WT and Hdh+/Q111 mice, we observed a differential m6A methylation in the distinct transcript regions (5´ and 3´UTR, exons and introns), which could induce such effects on downstream mRNA processing and metabolism in the HD condition. Supporting this possibility, we found several transcripts hypermethylated that have previously been described to be affected by mis-splicing or polyA changes [18, 21]. Majority of the m6A peaks of the methylated transcripts that overlap with mis-spliced genes are found in introns and exons, which are more likely to undergo alternative splicing as previously described [29]. Indeed, m6A modifications in introns are associated with long, slowly processed introns and alternative splicing events [67].
Our data also suggest a possible role of m6A in the activity-dependent alternative splicing processes required to achieve homeostatic control of neuronal output in health and diseased states. We observed under training conditions that most upregulated or downregulated peaks in WT and Hdh+/Q111 mice occur in introns. In the mammalian nervous system, alternative pre-mRNA splicing generates distinct functional isoforms that play key roles in normal physiology, supporting development, plasticity, complex behaviors, and cognition [94]. Strikingly, among the differential methylated genes in Hdh+/Q111 mice after training we identified Shank1 and Nrxn1. Shank 1, which plays an important role as scaffold protein in the formation of the PSD, contains diverse domains that appears to be regulated by alternative splicing [95]. Neurexin 1 (Nrxn1) also generates isoforms by alternative splicing enhancing postsynaptic NMDA-receptor-mediated synaptic responses [96]. All this evidence supports our hypothesis that m6A dysregulation could affect the fine tune of alternative splicing in Hdh+/Q111 mice, both under naive and trained conditions.
To understand the differential dynamic nature of the m6A RNA modifications observed in Hdh+/Q111 mice, we investigated the protein expression levels of FTO and METTL14 at different disease stages. In accordance with evidence showing correlation between abundance of METTL3 and METTL14 and learning efficiency in mouse hippocampus [39], we found increased nuclear METTL14 levels in WT mice after training. Importantly, this nuclear increase in the levels of METTL14 in response to neuronal activity demands during learning was not observed in Hdh+/Q111 mice and could be associated to the reduced methylation of transcripts observed after training. On the other hand, our data show that in WT mice after a memory-inducing activity, FTO levels rapidly increase in the nucleus and decrease in postsynaptic fractions. Within the nucleus, the observed increase of FTO levels in WT mice could clearly underlie synaptic remodeling by RNA processing as previous studies reported that overexpression of FTO results in increased co-localization with RNA processing factors [97]. On the contrary, memory-inducing activity is also known to rapidly decrease FTO in synapses increasing m6A to facilitate translation [35, 36, 76]. These local changes in FTO that are observed in response to behavioral experiences are also compatible with a role in regulation of synaptic plasticity in memory functions [75]. Remarkably, in Hdh+/Q111 mice FTO levels remain unchanged and steady FTO levels likely maintain a demethylation activity that would inhibit or disrupt translation or proper processing of synaptic genes which have already been described to be altered in HD [98,99,100].
Consistent with this dysregulation of FTO levels in Hdh+/Q111 mice, we detected an increased demethylation pattern in critical genes involved in memory processes and enriched in CA1 neurons. In accordance, FTO knockdown in CA1 region before training was sufficient to enhance spatial and recognition memory in Hdh+/Q111 mice. Interestingly, FTO knockdown improved the exploratory behavior in Hdh+/Q111 mice raising the question whether the enhanced memory observed by FTO knockdown was due to this increased exploratory behavior. It is known that processes like exploration, novelty and choice affects hippocampal function and memory via dopaminergic signaling which can be modulated by FTO [101,102,103,104,105]. Alterations in hippocampal dopaminergic signaling in HD mice has been reported to participate in hippocampal-dependent learning and memory deficits [106]. Hence, we hypothesize that FTO knockdown in CA1 neurons increased methylation of genes involved in the control of dopamine circuitry neurotransmission during training, promoting exploratory behavior and facilitating learning. Further analysis of FTO target genes in HD mice would provide better understanding of the signaling pathways affected by FTO dysregulation during memory formation in the affected HD brain.
The m6A dysregulation demonstrated in this study provides a missing signature that might define the aberrant post-transcriptional events underlying neuronal dysfunction in HD. Furthermore, our results open new avenues of investigation into the pathophysiological function of m6A during disease progression which might guide the development of new therapeutic strategies for HD.
Availability of supporting data
The sequence data, narrow Peak, and alignment data supporting the conclusions of this article are available in the NCBI GEO repository under the accession code GSE175618.
References
Foroud T, Siemers E, Kleindorfer D et al (1995) Cognitive scores in carriers of Huntington’s disease gene compared to noncarriers. Ann Neurol 37:657–664. https://doi.org/10.1002/ana.410370516
Lemiere J, Decruyenaere M, Evers-Kiebooms G et al (2004) Cognitive changes in patients with Huntington’s disease (HD) and asymptomatic carriers of the HD mutation. J Neurol. https://doi.org/10.1007/s00415-004-0461-9
Harris KL, Armstrong M, Swain R et al (2019) Huntington’s disease patients display progressive deficits in hippocampal-dependent cognition during a task of spatial memory. Cortex 119:417–427. https://doi.org/10.1016/j.cortex.2019.07.014
Giralt A, Saavedra A, Alberch J, Pérez-Navarro E (2012) Cognitive dysfunction in Huntington’s disease: humans, mouse models and molecular mechanisms. J Huntington’s Dis 1:155–173. https://doi.org/10.3233/JHD-120023
Ille R, Schäfer A, Scharmüller W et al (2011) Emotion recognition and experience in Huntington disease: a voxel-based morphometry study. J Psychiatry Neurosci 36:383–390. https://doi.org/10.1503/jpn.100143
Begeti F, Schwab LC, Mason SL, Barker RA (2016) Hippocampal dysfunction defines disease onset in Huntington’s disease. J Neurol Neurosurg Psychiatry 87:975–981. https://doi.org/10.1136/jnnp-2015-312413
Berrios GE, Wagle AC, Marková IS et al (2002) Psychiatric symptoms in neurologically asymptomatic Huntington’s disease gene carriers: a comparison with gene negative at risk subjects. Acta Psychiatr Scand 105:224–230. https://doi.org/10.1034/j.1600-0447.2002.0o456.x
Giralt A, Puigdellivol M, Carreton O et al (2012) Long-term memory deficits in Huntington’s disease are associated with reduced CBP histone acetylase activity. Hum Mol Genet 21:1203–1216. https://doi.org/10.1093/hmg/ddr552
Milnerwood AJ, Raymond LA (2007) Corticostriatal synaptic function in mouse models of Huntington’s disease: early effects of huntingtin repeat length and protein load. J Physiol 585:817–831. https://doi.org/10.1113/jphysiol.2007.142448
Spires TL, Grote HE, Garry S et al (2004) Dendritic spine pathology and deficits in experience-dependent dendritic plasticity in R6/1 Huntington’s disease transgenic mice. Eur J Neurosci 19:2799–2807. https://doi.org/10.1111/j.0953-816X.2004.03374.x
Usdin MT, Shelbourne PF, Myers RM, Madison DV (1999) Impaired synaptic plasticity in mice carrying the Huntington’s disease mutation. Hum Mol Genet 8:839–846. https://doi.org/10.1093/hmg/8.5.839
Lynch G, Kramar EA, Rex CS et al (2007) Brain-derived neurotrophic factor restores synaptic plasticity in a knock-in mouse model of Huntington’s disease. J Neurosci 27:4424–4434. https://doi.org/10.1523/JNEUROSCI.5113-06.2007
Brito V, Giralt A, Enriquez-Barreto L et al (2014) Neurotrophin receptor p75(NTR) mediates Huntington’s disease-associated synaptic and memory dysfunction. J Clin Invest 124:4411–4428. https://doi.org/10.1172/JCI74809
Yildirim F, Ng CW, Kappes V et al (2019) Early epigenomic and transcriptional changes reveal Elk-1 transcription factor as a therapeutic target in Huntington’s disease. Proc Natl Acad Sci 116:24840–24851. https://doi.org/10.1073/pnas.1908113116
Moumné L, Betuing S, Caboche J (2013) Multiple aspects of gene dysregulation in Huntington’s disease. Front Neurol. https://doi.org/10.3389/fneur.2013.00127
Suelves N, Kirkham-McCarthy L, Lahue RS, Ginés S (2017) A selective inhibitor of histone deacetylase 3 prevents cognitive deficits and suppresses striatal CAG repeat expansions in Huntington’s disease mice. Sci Rep 7:6082. https://doi.org/10.1038/s41598-017-05125-2
Romo L, Ashar-Patel A, Pfister E, Aronin N (2017) Alterations in mRNA 3′ UTR isoform abundance accompany gene expression changes in human Huntington’s disease brains. Cell Rep 20:3057–3070. https://doi.org/10.1016/j.celrep.2017.09.009
Elorza A, Márquez Y, Cabrera JR et al (2021) Huntington’s disease-specific mis-splicing unveils key effector genes and altered splicing factors. Brain 144:2009–2023. https://doi.org/10.1093/brain/awab087
Lin L, Park JW, Ramachandran S et al (2016) Transcriptome sequencing reveals aberrant alternative splicing in Huntington’s disease. Hum Mol Genet 25:3454–3466. https://doi.org/10.1093/hmg/ddw187
Lee S-T, Chu K, Im W-S et al (2011) Altered microRNA regulation in Huntington’s disease models. Exp Neurol 227:172–179. https://doi.org/10.1016/j.expneurol.2010.10.012
Picó S, Parras A, Santos-Galindo M et al (2021) CPEB alteration and aberrant transcriptome-polyadenylation lead to a treatable SLC19A3 deficiency in Huntington’s disease. Sci Transl Med. https://doi.org/10.1126/scitranslmed.abe7104
Chang M, Lv H, Zhang W et al (2017) Region-specific RNA m6 A methylation represents a new layer of control in the gene regulatory network in the mouse brain. Open Biol 7:170166. https://doi.org/10.1098/rsob.170166
Meyer KD, Saletore Y, Zumbo P et al (2012) Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149:1635–1646. https://doi.org/10.1016/j.cell.2012.05.003
Ping X-L, Sun B-F, Wang L et al (2014) Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res 24:177–189. https://doi.org/10.1038/cr.2014.3
Liu J, Yue Y, Han D et al (2014) A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 10:93–95. https://doi.org/10.1038/nchembio.1432
Zheng G, Dahl JA, Niu Y et al (2013) ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 49:18–29. https://doi.org/10.1016/j.molcel.2012.10.015
Jia G, Fu Y, Zhao X et al (2011) N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 7:885–887. https://doi.org/10.1038/nchembio.687
Shi H, Wei J, He C (2019) Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell 74:640–650. https://doi.org/10.1016/j.molcel.2019.04.025
Dominissini D, Moshitch-Moshkovitz S, Schwartz S et al (2012) Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485:201–206. https://doi.org/10.1038/nature11112
Zhao X, Yang Y, Sun B-F et al (2014) FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res 24:1403–1419. https://doi.org/10.1038/cr.2014.151
Xiao W, Adhikari S, Dahal U et al (2016) Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol Cell 61:507–519. https://doi.org/10.1016/j.molcel.2016.01.012
Wang X, Zhao BS, Roundtree IA et al (2015) N6-methyladenosine modulates messenger RNA translation efficiency. Cell 161:1388–1399. https://doi.org/10.1016/j.cell.2015.05.014
Roundtree IA, Evans ME, Pan T, He C (2017) Dynamic RNA modifications in gene expression regulation. Cell 169:1187–1200. https://doi.org/10.1016/j.cell.2017.05.045
Zhou KI, Shi H, Lyu R et al (2019) Regulation of co-transcriptional pre-mRNA splicing by m6A through the low-complexity protein hnRNPG. Mol Cell 76:70-81.e9. https://doi.org/10.1016/j.molcel.2019.07.005
Widagdo J, Zhao Q-Y, Kempen M-J et al (2016) Experience-dependent accumulation of N6-methyladenosine in the prefrontal cortex is associated with memory processes in mice. J Neurosci 36:6771–6777. https://doi.org/10.1523/JNEUROSCI.4053-15.2016
Walters BJ, Mercaldo V, Gillon CJ et al (2017) The role of the RNA demethylase FTO (fat mass and obesity-associated) and mRNA methylation in hippocampal memory formation. Neuropsychopharmacology 42:1502–1510. https://doi.org/10.1038/npp.2017.31
Koranda JL, Dore L, Shi H et al (2018) Mettl14 is essential for epitranscriptomic regulation of striatal function and learning. Neuron 99:283-292.e5. https://doi.org/10.1016/j.neuron.2018.06.007
Shi H, Zhang X, Weng Y-L et al (2018) m6A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature 563:249–253. https://doi.org/10.1038/s41586-018-0666-1
Zhang Z, Wang M, Xie D et al (2018) METTL3-mediated N6-methyladenosine mRNA modification enhances long-term memory consolidation. Cell Res 28:1050–1061. https://doi.org/10.1038/s41422-018-0092-9
Merkurjev D, Hong W-T, Iida K et al (2018) Synaptic N6-methyladenosine (m6A) epitranscriptome reveals functional partitioning of localized transcripts. Nat Neurosci 21:1004–1014. https://doi.org/10.1038/s41593-018-0173-6
Nussbacher JK, Tabet R, Yeo GW, Lagier-Tourenne C (2019) Disruption of RNA metabolism in neurological diseases and emerging therapeutic interventions. Neuron 102:294–320. https://doi.org/10.1016/j.neuron.2019.03.014
Zhao F, Xu Y, Gao S et al (2021) METTL3-dependent RNA m6A dysregulation contributes to neurodegeneration in Alzheimer’s disease through aberrant cell cycle events. Mol Neurodegener 16:70. https://doi.org/10.1186/s13024-021-00484-x
Huang H, Camats-Perna J, Medeiros R et al (2020) Altered expression of the m6A methyltransferase METTL3 in Alzheimer’s disease. Eneuro. https://doi.org/10.1523/ENEURO.0125-20.2020
Han M, Liu Z, Xu Y et al (2020) Abnormality of m6A mRNA methylation is involved in Alzheimer’s disease. Front Neurosci. https://doi.org/10.3389/fnins.2020.00098
Chen X, Yu C, Guo M et al (2019) Down-regulation of m6A mRNA methylation is involved in dopaminergic neuronal death. ACS Chem Neurosci 10:2355–2363. https://doi.org/10.1021/acschemneuro.8b00657
Qin L, Min S, Shu L et al (2020) Genetic analysis of N6-methyladenosine modification genes in Parkinson’s disease. Neurobiol Aging 93:143.e9-143.e13. https://doi.org/10.1016/j.neurobiolaging.2020.03.018
Puigdellívol M, Cherubini M, Brito V et al (2015) A role for Kalirin-7 in corticostriatal synaptic dysfunction in Huntington’s disease. Hum Mol Genet 24:7265–7285. https://doi.org/10.1093/hmg/ddv426
Wheeler V (1999) Length-dependent gametic CAG repeat instability in the Huntington’s disease knock-in mouse. Hum Mol Genet 8:115–122. https://doi.org/10.1093/hmg/8.1.115
Mendel M, Delaney K, Pandey RR et al (2021) Splice site m6A methylation prevents binding of U2AF35 to inhibit RNA splicing. Cell 184:3125-3142.e25. https://doi.org/10.1016/j.cell.2021.03.062
Zeng Y, Wang S, Gao S et al (2018) Refined RIP-seq protocol for epitranscriptome analysis with low input materials. PLoS Biol 16:e2006092. https://doi.org/10.1371/journal.pbio.2006092
Dobin A, Davis CA, Schlesinger F et al (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. https://doi.org/10.1093/bioinformatics/bts635
Li H, Handsaker B, Wysoker A et al (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. https://doi.org/10.1093/bioinformatics/btp352
Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. https://doi.org/10.1093/bioinformatics/btq033
Zhang Y, Liu T, Meyer CA et al (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9:R137. https://doi.org/10.1186/gb-2008-9-9-r137
Li Q, Brown JB, Huang H, Bickel PJ (2011) Measuring reproducibility of high-throughput experiments. Ann Appl Stat. https://doi.org/10.1214/11-AOAS466
Ross-Innes CS, Stark R, Teschendorff AE et al (2012) Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481:389–393. https://doi.org/10.1038/nature10730
Yu G, Wang L-G, He Q-Y (2015) ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31:2382–2383. https://doi.org/10.1093/bioinformatics/btv145
Koopmans F, van Nierop P, Andres-Alonso M et al (2019) SynGO: an evidence-based, expert-curated knowledge base for the synapse. Neuron 103:217-234.e4. https://doi.org/10.1016/j.neuron.2019.05.002
Heinz S, Benner C, Spann N et al (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38:576–589. https://doi.org/10.1016/j.molcel.2010.05.004
Law CW, Alhamdoosh M, Su S et al (2018) RNA-seq analysis is easy as 1–2–3 with limma, Glimma and edgeR. FRes 5:1408. https://doi.org/10.12688/f1000research.9005.3
McCarthy DJ, Chen Y, Smyth GK (2012) Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40:4288–4297. https://doi.org/10.1093/nar/gks042
Law CW, Chen Y, Shi W, Smyth GK (2014) voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15:R29. https://doi.org/10.1186/gb-2014-15-2-r29
Bruyère J, Abada Y-S, Vitet H et al (2020) Presynaptic APP levels and synaptic homeostasis are regulated by Akt phosphorylation of huntingtin. Elife. https://doi.org/10.7554/eLife.56371
Du H, Zhao Y, He J et al (2016) YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4–NOT deadenylase complex. Nat Commun 7:12626. https://doi.org/10.1038/ncomms12626
Yue Y, Liu J, Cui X et al (2018) VIRMA mediates preferential m6A mRNA methylation in 3′UTR and near stop codon and associates with alternative polyadenylation. Cell Dis 4:10. https://doi.org/10.1038/s41421-018-0019-0
Xiao W, Adhikari S, Dahal U et al (2016) Nuclear m 6 A reader YTHDC1 regulates mRNA splicing. Mol Cell 61:507–519. https://doi.org/10.1016/j.molcel.2016.01.012
Louloupi A, Ntini E, Conrad T, Ørom UAV (2018) Transient N-6-methyladenosine transcriptome sequencing reveals a regulatory role of m6A in splicing efficiency. Cell Rep 23:3429–3437. https://doi.org/10.1016/j.celrep.2018.05.077
Tabrizi SJ, Scahill RI, Owen G et al (2013) Predictors of phenotypic progression and disease onset in premanifest and early-stage Huntington’s disease in the TRACK-HD study: analysis of 36-month observational data. Lancet Neurol 12:637–649. https://doi.org/10.1016/S1474-4422(13)70088-7
Butters N, Wolfe J, Martone M et al (1985) Memory disorders associated with Huntington’s disease: verbal recall, verbal recognition and procedural memory. Neuropsychologia 23:729–743. https://doi.org/10.1016/0028-3932(85)90080-6
Nguyen HP, Kobbe P, Rahne H et al (2006) Behavioral abnormalities precede neuropathological markers in rats transgenic for Huntington’s disease. Hum Mol Genet 15:3177–3194. https://doi.org/10.1093/hmg/ddl394
Bolivar VJ, Manley K, Messer A (2004) Early exploratory behavior abnormalities in R6/1 Huntington’s disease transgenic mice. Brain Res 1005:29–35. https://doi.org/10.1016/j.brainres.2004.01.021
Paulsen JS, Langbehn DR, Stout JC et al (2008) Detection of Huntington’s disease decades before diagnosis: the Predict-HD study. J Neurol Neurosurg Psychiatry 79:874–880. https://doi.org/10.1136/jnnp.2007.128728
Tsien JZ, Huerta PT, Tonegawa S (1996) The essential role of hippocampal CA1 NMDA receptor-dependent synaptic plasticity in spatial memory. Cell 87:1327–1338. https://doi.org/10.1016/S0092-8674(00)81827-9
Halder R, Hennion M, Vidal RO et al (2016) DNA methylation changes in plasticity genes accompany the formation and maintenance of memory. Nat Neurosci 19:102–110. https://doi.org/10.1038/nn.4194
Livneh I, Moshitch-Moshkovitz S, Amariglio N et al (2020) The m6A epitranscriptome: transcriptome plasticity in brain development and function. Nat Rev Neurosci 21:36–51. https://doi.org/10.1038/s41583-019-0244-z
Engel M, Eggert C, Kaplick PM et al (2018) The role of m6A/m-RNA methylation in stress response regulation. Neuron 99:389-403.e9. https://doi.org/10.1016/j.neuron.2018.07.009
Murai T, Okuda S, Tanaka T, Ohta H (2007) Characteristics of object location memory in mice: behavioral and pharmacological studies. Physiol Behav 90:116–124. https://doi.org/10.1016/j.physbeh.2006.09.013
Assini FL, Duzzioni M, Takahashi RN (2009) Object location memory in mice: pharmacological validation and further evidence of hippocampal CA1 participation. Behav Brain Res 204:206–211. https://doi.org/10.1016/j.bbr.2009.06.005
Antunes M, Biala G (2012) The novel object recognition memory: neurobiology, test procedure, and its modifications. Cogn Process 13:93–110. https://doi.org/10.1007/s10339-011-0430-z
Niccolini F, Haider S, Reis Marques T et al (2015) Altered PDE10A expression detectable early before symptomatic onset in Huntington’s disease. Brain 138:3016–3029. https://doi.org/10.1093/brain/awv214
Hu H, McCaw EA, Hebb ALO et al (2004) Mutant huntingtin affects the rate of transcription of striatum-specific isoforms of phosphodiesterase 10A. Eur J Neurosci 20:3351–3363. https://doi.org/10.1111/j.1460-9568.2004.03796.x
Borovecki F, Lovrecic L, Zhou J et al (2005) Genome-wide expression profiling of human blood reveals biomarkers for Huntington’s disease. Proc Natl Acad Sci 102:11023–11028. https://doi.org/10.1073/pnas.0504921102
Brito V, Puigdellívol M, Giralt A et al (2013) Imbalance of p75NTR/TrkB protein expression in Huntington’s disease: implication for neuroprotective therapies. Cell Death Dis 4:e595–e595. https://doi.org/10.1038/cddis.2013.116
Saft C, Epplen JT, Wieczorek S et al (2011) NMDA receptor gene variations as modifiers in Huntington disease: a replication study. PLoS Curr 3:RRN1247. https://doi.org/10.1371/currents.RRN1247
Arning L, Saft C, Wieczorek S et al (2007) NR2A and NR2B receptor gene variations modify age at onset in Huntington disease in a sex-specific manner. Hum Genet 122:175–182. https://doi.org/10.1007/s00439-007-0393-4
Kang JE, Choi SA, Park JB, Chung KC (2005) Regulation of the proapoptotic activity of huntingtin interacting protein 1 by Dyrk1 and caspase-3 in hippocampal neuroprogenitor cells. J Neurosci Res 81:62–72. https://doi.org/10.1002/jnr.20534
Francelle L, Lotz C, Outeiro T et al (2017) Contribution of neuroepigenetics to Huntington’s disease. Front Hum Neurosci. https://doi.org/10.3389/fnhum.2017.00017
Kumar A, Vaish M, Ratan RR (2014) Transcriptional dysregulation in Huntington’s disease: a failure of adaptive transcriptional homeostasis. Drug Dis Today 19:956–962. https://doi.org/10.1016/j.drudis.2014.03.016
Martinez De La Cruz B, Markus R, Malla S et al (2021) Modifying the m6A brain methylome by ALKBH5-mediated demethylation: a new contender for synaptic tagging. Mol Psychiatry. https://doi.org/10.1038/s41380-021-01282-z
Xuan J-J, Sun W-J, Lin P-H et al (2018) RMBase v2.0: deciphering the map of RNA modifications from epitranscriptome sequencing data. Nucleic Acids Res 46:D327–D334. https://doi.org/10.1093/nar/gkx934
Neueder A, Landles C, Ghosh R et al (2017) The pathogenic exon 1 HTT protein is produced by incomplete splicing in Huntington’s disease patients. Sci Rep 7:1307. https://doi.org/10.1038/s41598-017-01510-z
Slobodin B, Han R, Calderone V et al (2017) Transcription impacts the efficiency of mRNA translation via co-transcriptional N6-adenosine methylation. Cell 169:326-337.e12. https://doi.org/10.1016/j.cell.2017.03.031
Kasowitz SD, Ma J, Anderson SJ et al (2018) Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genet 14:e1007412. https://doi.org/10.1371/journal.pgen.1007412
Lopez Soto EJ, Gandal MJ, Gonatopoulos-Pournatzis T et al (2019) Mechanisms of neuronal alternative splicing and strategies for therapeutic interventions. J Neurosci 39:8193–8199. https://doi.org/10.1523/jneurosci.1149-19.2019
Lim S, Naisbitt S, Yoon J et al (1999) Characterization of the Shank family of synaptic proteins. J Biol Chem 274:29510–29518. https://doi.org/10.1074/jbc.274.41.29510
Dai J, Aoto J, Südhof TC (2019) Alternative splicing of presynaptic neurexins differentially controls postsynaptic NMDA and AMPA receptor responses. Neuron 102:993-1008.e5. https://doi.org/10.1016/j.neuron.2019.03.032
Berulava T, Horsthemke B (2010) The obesity-associated SNPs in intron 1 of the FTO gene affect primary transcript levels. Eur J Hum Genet 18:1054–1056. https://doi.org/10.1038/ejhg.2010.71
Creus-Muncunill J, Badillos-Rodríguez R, Garcia-Forn M et al (2019) Increased translation as a novel pathogenic mechanism in Huntington’s disease. Brain 142:3158–3175. https://doi.org/10.1093/brain/awz230
Eshraghi M, Karunadharma PP, Blin J et al (2021) Mutant Huntingtin stalls ribosomes and represses protein synthesis in a cellular model of Huntington disease. Nat Commun 12:1461. https://doi.org/10.1038/s41467-021-21637-y
Barron JC, Hurley EP, Parsons MP (2021) Huntingtin and the synapse. Front Cell Neurosci. https://doi.org/10.3389/fncel.2021.689332
Düzel E, Bunzeck N, Guitart-Masip M, Düzel S (2010) Novelty-related motivation of anticipation and exploration by dopamine (NOMAD): implications for healthy aging. Neurosci Biobehav Rev 34:660–669. https://doi.org/10.1016/j.neubiorev.2009.08.006
Lisman JE, Grace AA (2005) The hippocampal-VTA loop: controlling the entry of information into long-term memory. Neuron 46:703–713. https://doi.org/10.1016/j.neuron.2005.05.002
Murty VP, DuBrow S, Davachi L (2015) The simple act of choosing influences declarative memory. J Neurosci 35:6255–6264. https://doi.org/10.1523/JNEUROSCI.4181-14.2015
Hess ME, Hess S, Meyer KD et al (2013) The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat Neurosci 16:1042–1048. https://doi.org/10.1038/nn.3449
Ruud J, Alber J, Tokarska A et al (2019) The fat mass and obesity-associated protein (FTO) regulates locomotor responses to novelty via D2R medium spiny neurons. Cell Rep 27:3182-3198.e9. https://doi.org/10.1016/j.celrep.2019.05.037
Tyebji S, Saavedra A, Canas PM et al (2015) Hyperactivation of D1 and A2A receptors contributes to cognitive dysfunction in Huntington’s disease. Neurobiol Dis 74:41–57. https://doi.org/10.1016/j.nbd.2014.11.004
Acknowledgements
We are very grateful to Ana Lopez and Maria Teresa Muñoz for technical assistance, Dr Teresa Rodrigo and the staff of the animal care facility (Facultat de Psicologia Universitat de Barcelona), and Dr. Maria Calvo from the Advanced Optical Microscopy Unit from Scientific and Technological Centers from University of Barcelona for their support and advice with confocal technique. We thank members of our laboratory for helpful discussion.
Funding
Open Access funding provided thanks to the CRUE-CSIC agreement with Springer Nature. This work was supported by Ministerio de Economía y Competitividad (PID2020-116474RB-100 to V.B. and RTI2018-094374-B100 to S.G.); Hereditary Disease Foundation grant to V.B.; Centro de Investigaciones Biomédicas en Red sobre Enfermedades Neurodegenerativas (CIBERNED) to S.G.; PhD grant program by La Generalitat de Catalunya (2018FI_B_00487) to A.P. Ørom lab was supported by Novo Nordisk Foundation, Lundbeck Foundation, Independent Research Fund Denmark, Danish Cancer Society and Carlsbergfonden.
Author information
Authors and Affiliations
Contributions
AP carried out most experiments, analyzed and interpreted results and wrote the manuscript. AS performed the MeRIP-seq data analysis and differential expression analysis, ASB performed surgeries and behavioral experiments. ME participated in collection of samples. RAV, EM and AG analyzed and interpreted results. UVO supervised MeRIP-seq experiments, analyzed and interpreted data. SG and VB conceived and supervised the study, analyzed results, and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Study approval
All procedures were performed in compliance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and approved by the local animal care committee of the Universitat de Barcelona (448/17) and the Generalitat de Catalunya (9878 P2), in accordance with the European (2010/63/EU) and Spanish (RD53/2013) guidelines for the care and use of laboratory animals.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pupak, A., Singh, A., Sancho-Balsells, A. et al. Altered m6A RNA methylation contributes to hippocampal memory deficits in Huntington’s disease mice. Cell. Mol. Life Sci. 79, 416 (2022). https://doi.org/10.1007/s00018-022-04444-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-022-04444-6