Abstract
Background
Janus kinase 3 (JAK3) acts downstream of the interleukin-2 (IL-2) receptor family to play a pivotal role in the regulation of lymphoid cell development. Activating JAK3 mutations are associated with a number of lymphoid and other malignancies, with mutations within the regulatory pseudokinase domain common.
Methods
The pseudokinase domain mutations A572V and A573V were separately introduced into the highly conserved zebrafish Jak3 and transiently expressed in cell lines and zebrafish embryos to examine their activity and impact on early T cells. Genome editing was subsequently used to introduce the A573V mutation into the zebrafish genome to study the effects of JAK3 activation on lymphoid cells in a physiologically relevant context throughout the life-course.
Results
Zebrafish Jak3 A573V produced the strongest activation of downstream STAT5 in vitro and elicited a significant increase in T cells in zebrafish embryos. Zebrafish carrying just a single copy of the Jak3 A573V allele displayed elevated embryonic T cells, which continued into adulthood. Hematopoietic precursors and NK cells were also increased, but not B cells. The lymphoproliferative effects of Jak3 A573V in embryos was shown to be dependent on zebrafish IL-2Rγc, JAK1 and STAT5B equivalents, and could be suppressed with the JAK3 inhibitor Tofacitinib.
Conclusions
This study demonstrates that a single JAK3 A573V allele expressed from the endogenous locus was able to enhance lymphopoiesis throughout the life-course, which was mediated via an IL-2Rγc/JAK1/JAK3/STAT5 signaling pathway and was sensitive to Tofacitinib. This extends our understanding of oncogenic JAK3 mutations and creates a novel model to underpin further translational investigations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Cytokine receptor signaling via the Janus kinase-Signal transducer and activator of transcription (JAK-STAT) pathway represents a core mode of regulation in the context of blood and immune cell development and function [50, 58]. Of the four mammalian JAKs, JAK3 is unique in having restricted expression in hematopoietic cells, particularly within the lymphoid compartment [18]. JAK3 exclusively associates with the common interleukin-2 receptor gamma common (IL-2Rγc) chain that is shared by the receptors of several cytokines critical for lymphopoiesis, specifically IL-2, IL-4, IL-7, IL-9, IL-15 and IL-21 [57]. JAK1 has been shown to be critical for signaling from these receptors [20, 54], which occurs via several intracellular signaling pathways, but particularly STAT5 [34, 43].
The importance of JAK3 in normal lymphopoiesis is illustrated by inactivating mutations that cause autosomal Severe Combined Immune Deficiency (SCID) that is characterized by reduced T and NK cells as well as non-functional B cells, mediated by loss of function of the suite of IL-2Rγc-utilizing receptors [10]. In contrast, constitutively activating JAK3 mutations—variously impacting the FERM, SH2, pseudokinase and kinase domains—have been detected in in several cases of human hematopoietic neoplasms, specifically cutaneous T cell lymphoma (CTCL) [11, 45], intestinal T cell lymphoma [49], T cell acute lymphoblastic leukemia (T-ALL) [3, 48], T cell prolymphocytic leukemia (T-PLL) [6], Natural killer T cell lymphoma (NKTCL) [27], as well as acute megakaryoblastic leukemia (AMKL) [25, 56, 64]. Two common mutations within the pseudokinase domain (PKD), A572V and A573V, have been identified in NKTCL [8, 27], T-ALL [3, 17], intestinal T cell lymphoma [49], T-PLL [6] and AMKL [44, 64]. These mutations cause JAK3 to be constitutively activated and able to mediate ligand-independent proliferation of cells expressing them, including a range of lymphoid cells [27, 64]. JAK1 has been shown to be essential for JAK3 mutants to mediate their effects [17, 54].
Zebrafish show strong conservation of cytokine receptor signaling through the JAK-STAT pathway [37, 38], including a conserved role for the IL-2Rγc in immune cell development [59, 60]. This study examined the A572V and A573V JAK3 PKD mutations, which were introduced into zebrafish Jak3 and the effects on STAT5 activation in vitro and T lymphocyte production in vivo analyzed. A mutant zebrafish jak3 allele encoding the Jak3 A573V mutant was generated by genome editing, with fish carrying one or two alleles analyzed for immune cell development from embryonic through to adult stages. Genetic analysis was used to assess the importance of IL-2Rγc, JAK1 and STAT5 in mediating the effects of the JAK3 A573V mutation, and sensitivity to the JAK3 inhibitor Tofacitinib analyzed. Collectively, this work provides critical in vivo insights into oncogenic JAK3.
Methods
In vitro analyses
Human HEK293T cells were transfected with pBKCMV expressing HA-tagged zebrafish Jak3 wild type [60] as well as A572V and A573V mutants (synthesized by Genewiz LLC), along with zebrafish Stat5.1 [32] and analyzed by Western blot with anti-phospho-STAT5, anti-STAT5 and anti-GAPDH, as described [60].
Fish husbandry and genetic manipulations
Zebrafish were maintained using standard husbandry practices [65], following national guidelines for their care and use, with all studies approved by the Deakin University Animal Ethics Committee. Wild-type embryos at the 1 cell stage were injected with 100 pg/μl in vitro transcribed, capped mRNA encoding zebrafish Jak3 A572V and A573V mutants. For CRISPR/Cas9 genome editing, a guide RNA that targeted exon 13 of the jak3 gene was produced using a specific primer pair (5′-TAGGAGATTTGACTCATCAAAC and 5′-AAACGTTTGATGAGTCAAATCT), as described [21]. Wild-type embryos were injected with 12.5 ng/μl gRNA, 100 ng/μl Cas9 mRNA (Sigma) and 10 μM homology-dependent repair (HDR) oligonucleotide (5’-TCTTCTTTTTTCTAGTCTCTTTTCGAGGCGGTATCCTTGATGAGTCAAATCTCCCACAGGCACCTTC), raised to adulthood and outcrossed with wild-type fish. Carriers of the jak3 mutant allele were identified and, following an additional round of outcrossing, were in-crossed to generate wild-type, heterozygote and homozygote mutant progeny for analysis. The jak3 mutant was also crossed onto lines carrying hypomorphic mutations in il2rga [59], as well as jak1, stat5.1 and stat5.2 produced in-house (unpublished data) to generate additional genotypes for analysis.
Genomic DNA analysis
Genomic DNA from adult fin clips and whole embryos was isolated with QuickExtract following the manufacturer’s instructions. This was subjected to PCR with jak3-specific primers, either for restriction fragment length polymorphism (RFLP) analysis with BciVI (5′-TTATCCATGTGAATAAATGTTTAATCTTC, 5′-CTAATGCCATACACCAAAAGAAGG) or High Resolution Melt (HRM) (5′-TTATCCATGTGAATAAATGTTTAATCTTC, 5′-CTAATGCCATACACCAAAAGAAGG) using Precision Melt Suremix and Analysis Software (BioRad) to identify jak3 mutants. These were individually confirmed by Sanger sequencing at the Australian Genome Research Facility. Genotyping details for the other gene mutations is included in the relevant Supplementary Figures.
RT-PCR and qRT2-PCR
Total RNA was extracted from individual 28 dpf juvenile zebrafish with RNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol for animal tissues. This was subjected to semi-quantitative reverse-transcription polymerase chain reaction (RT-PCR) with primers for T-cell receptor beta (TCRβ) variable chains vb1.5/17.5 (5′-AATGGACAGCTTGATAGAACTGAAC, 5′-TGCTTATTCAACCGAACAGAAACATTC), vb12 (5′-CAGACACCGTGCTTCAGTCGAG, 5′-ACGTTTCATGGCAGTGTTACCTG) and vb14.5 (5′-GAATCCAATGTGACGTTAACATGC, 5′-CATGATCATAAGGACCACTACAG) and immunoglobulin variable heavy chains igvh1 (5′-GATGGACGTGTTACAATTTGG, 5′-CCTCCTCAGACTCTGTGGTGA) and igvh4 (5′-CAAGATGAAGAATGCTCTCTG, 5′-TGTCAAAGTATGGAGTCGA) or quantitative real-time RT-PCR (qRT2-PCR) with actb (5′-TGGCATCACACCTTCTAC, 5′-AGACCATCACCAGAGTCC), cmyb (5′-TCGGCAAGACACGCTGGA, 5′-AATGCTTTGCGATTACTGACCA), rag1 (5′-GGATGTGAAGTATGTGTGTTTGA, 5′-TGGAACCCAGGGAGAAGC), cd4 (5′-TCTTGCTTGTTGCATTCGCC, 5′-TCCCTTTGGCTGTTTGTTATTGT), cd8 (5′-ACTCTTCTTCGGAGAGGTGAC, 5-ACAGGCTTCAGTGTTGTTTGAA), cd79a (5′-GCGAGGGTGTGAAAAACAGT, 5-CCCTTTCTGTCTTCCTGTCCA), igm (5′-CCGAATACAGTGCCACAAGC, 5′-TCTCCCTGCTATCTTTCCGC), nccrp1 (5′-TCAGCACAGGTGGTTCACTCTA, 5′-GGCTTTCCTCATACCAGTCTTC) and nkld (5′-TGGTGAAATCCCAACAGAGCA, 5′-TTTCATCCTGAGTTGCACCA). Data were normalized to actb and fold change was calculated using the ΔΔCt method.
Whole-mount in situ hybridization (WISH) and hemoglobin staining
Anesthetized embryos were dechorionated and fixed in 4% (w/v) paraformaldehyde at 4°C before WISH with anti-sense digoxigenin-labeled gene-specific probes, as described (Thisse and [62], or subjected to staining of hemoglobin with O-dianisidine [35]. Quantitation was achieved by enumeration of individual cells or measuring the area of staining relative to eye diameter using CellSens Dimension 1.6 software (Olympus) in a blind fashion on images taken on an Olympus MVX10 monozoom microscope with a 1 × MVXPlan Apochromat lens (NA = 0.25) with an Olympus DP72 camera. Data from approximately 30 embryos were analyzed for significance with a Student’s t test using Welch's correction where necessary, with data tested for normality.
Ex vivo analyses
Cytospin preparations were prepared from embryonic and adult blood as well as adult kidney and stained with Giemsa (Sigma), and differential counts performed. These were imaged on a Leica DM E microscope with a 100 × oil objective (NA = 1.25) with an Olympus SC50 camera Alternatively, adult zebrafish kidney cells were prepared in ice-cold phosphate-buffered saline supplemented with 2 mM EDTA and 2% (v/v) fetal calf serum and passaged through a 40 μm sieve and analyzed using a FACSCantoII with cell populations identified in a side-scatter (SSC)/forward-scatter (FSC) plot as described [4]. Data were analyzed for significance with a Student’s t test.
Survival analysis
Survival of juvenile/adult fish was monitored by regular visual inspection and displayed as a Kaplan–Meier curve with statistical significance determined using a log-rank (Mantel–Cox) test.
Results
Analysis of zebrafish Jak3 mutants
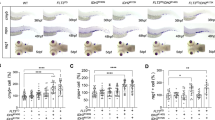
Zebrafish possess a highly conserved Jak3 [37] that contains the same domain structure as human JAK3 (Fig. 1A), and high amino acid identity extending to residues within the PKD, including those encompassing A572 and A573 (Fig. 1B). To analyze the effect of A572V and A573V mutations in the context of zebrafish Jak3, expression constructs encoding these variants and wild-type Jak3 tagged with HA were transfected into human HEK293 cells along with one encoding zebrafish Stat5.1, with Stat5.1 activation determined by Western blot using an anti-phospho-STAT5 antibody (Fig. 1C). Modest phospho-STAT5 was observed with Jak3 wild-type, as described [60], which was increased with Jak3 A572V and even more so with Jak3 A573V. Analysis with anti-STAT5, anti-HA and anti-GAPDH antibodies confirmed equivalent transfection and loading in each case (Fig. 1C). To confirm these effects in vivo, one-cell stage wild-type zebrafish embryos were injected with in vitro transcribed mRNA encoding Jak3 A572V or A573V and at 5 days post-fertilization (dpf) were subjected to WISH with rag1 as a marker of mature T lymphocytes [67]. This revealed increased expression of rag1 in embryos injected with Jak3 A573V, but not Jak3 A572V, compared to control uninjected embryos (Fig. 1D–G).
Conservation of human JAK3 constitutively-activating mutations in zebrafish. A JAK3 structure. Schematic representation of JAK3 including the FERM (pink), SH2 (green), pseudokinase (red) and kinase (blue) domains, which are all present in zebrafish Jak3. B Conservation of human and zebrafish JAK3. Human JAK3 and zebrafish Jak3 were aligned using CLUSTALX, showing identical (*) and highly similar (:) residues around A572/A573. C Analysis of zebrafish Jak3 mutants in vitro. Western blot analysis of HEK293 cells transfected with wild-type Jak3 + Stat5.1 (WT), Jak3 A572V + Stat5.1 (A572V) and Jak3 A573V + Stat5.1 (A573V) using anti-phospho-STAT5 (pSTAT5), anti-STAT5, anti-HA (JAK3) and anti-GAPDH antibodies, as indicated. D–G Analysis of zebrafish Jak3 mutants in vivo. Uninjected (Uninj) embryos (D) or those injected with mRNA encoding Jak3 A572V (E) or A573V (F), were subjected to WISH with rag1 and imaged, with representative embryos shown. The area of rag1 expression was determined for individual embryos (G) with mean and SEM shown in red and level of statistical significance indicated (*p < 0.05, ns: not significant; n = 30) (6.3 × magnification, scale bar = 100 μm). H Genome targeting of zebrafish jak3. Exon–intron structure of the target site with spanning primers (F, R) indicated by black arrows, with exons encoding the pseudokinase domain shown as numbered boxes and introns represented as solid lines, with the Jak3 A573V mutation denoted by a purple arrow. I The jak3 constitutively activating (ca) mutant alleles generated in zebrafish. The nucleotide sequence of homozygote wild-type (+/+), along with heterozygote (+/ca) and homozygote (ca/ca) mutant zebrafish are shown with the five bases mutated indicated and the introduced BciVI site boxed. The protein translations are shown below in black text, with the exception of wild-type A573 in blue and mutant V573 in purple
Generation of zebrafish carrying T cell lymphoma-derived Jak3 mutation
Since the zebrafish Jak3 A573V mutant elicited robust Stat5.1 activation in vitro and increased T cells in vivo, it was selected for further study. To introduce this mutation in the zebrafish genome, a CRISPR/Cas9 HDR strategy was used [61]. Guide RNA targeting exon 13 was co-injected with Cas9-encoding mRNA along with a 67 bp single-stranded oligonucleotide repair template that contained missense nucleotide changes in the coding sequence to elicit a A573V mutation, as well as silent changes to introduce a restriction enzyme site (BciVI) to facilitate screening and destroy the protospacer adjacent motif to prevent further recognition by Cas9 (Fig. 1H–I). Injected embryos were raised to adulthood for screening, which identified zebrafish carrying an allele encoding the constitutively activating Jak3 A573V mutant (designated ca). These were outcrossed for two generations to remove any potential off-target mutations, followed by an in-cross to generate heterozygote Jak3 A573V (+/ca) mutants, mimicking patient mutations, as well as homozygote (ca/ca) mutants, which were confirmed by sequencing (Fig. 1I).
Impact of Jak3 A573V mutation on embryonic lymphoid cell development
Lymphocyte progenitors first populate the zebrafish thymus from approximately 2.5 dpf to initiate T lymphopoiesis [29]. To investigate the effect of the Jak3 A573V mutation on embryonic lymphopoiesis, wild type (jak3+/+) along with heterozygote (jak3+/ca), and homozygote (jak3ca/ca) mutants, were subjected to WISH with the early lymphoid marker ikzf1 [66], as well as the mature lymphocyte markers rag1 [67], lck [28] and tcra [13]. Both jak3+/ca and jak3ca/ca embryos showed a significant increase in ikzf1 expression compared to jak3+/+ embryos at both 3.5 dpf (Fig. 2A–D) and 5 dpf (Fig. 2E–H), with a similar increase also evident in the expression of rag1 (Fig. 2I–L), lck (Fig. 2M-P) and tcra (Fig. 2Q–T) at 5 dpf. Interestingly, no significant differences were observed between jak3+/ca and jak3ca/ca embryos (Fig. 2D, H, L, P, T). In contrast, the number of lyz+ leucocytes [39] (Fig. 2U–X), mpo+ neutrophils [35] (Fig. 2Y–B′) and the extent of O-dianisidine staining of erythrocytes [35] (Fig. 2C′–E′) was not significantly different between jak3+/+, jak3+/ca and jak3ca/ca embryos. Analysis of blood smears at 5 dpf confirmed a significant increase in the proportion of lymphocytes in jak3+/ca and jak3ca/ca embryos in comparison to their jak3+/+ siblings, but again no statistically significant difference between jak3+/ca and jak3ca/ca embryos was observed (Fig. 2F′–I′).
Phenotypic analysis of zebrafish carrying the Jak3 A573V encoding allele. Embryos homozygous for Jak3 wildtype (jak3+/+) or Jak3 A573V (jak3ca/ca) or those heterozygous for Jak3 A573V (jak3+/ca) were subjected to WISH with ikzf1 at 3.5 dpf (A–C) and 5 dpf (E–G), rag1 (I–K), lck (M–O), tcra (Q–S), lyz (U–W) and mpo (Y–A′) at 5 dpf, as well as O-dianisidine staining (C′–E′) (6.3/3.2 × magnification, scale bar = 200 μm) or their blood analyzed with Giemsa staining at 5 dpf (F′–H′; e, erythrocyte, l, lymphocyte) (100 × magnification, scale bar = 10 μm). Individual embryos were assessed for relative area of staining with ikzf1 (D, H), rag1 (L), lck (P) and tcrα (T), for individual embryos or total number of lyz+ +(X) and mpo+ (B′) cells, or blood differential counts (I′) with the mean and SEM shown in red and level of statistical significance indicated (*p < 0.05, **p < 0.01, ***p < 0.001, ns: not significant) (n = 30 for WISH analysis and n = 5 for blood counts)
Analysis of zebrafish harboring the Jak3 A573V mutation into adulthood
Zebrafish B lymphocytes develop from around 3 weeks post-fertilization [51], with NK-related cells also readily detectable at this time [46]. To examine the effects of Jak3 A573V mutation on these lineages, qRT2-PCR was performed on 28 dpf jak3+/+, jak3+/ca and jak3ca/ca larvae using primers specific for genes marking HSCs (cmyb), T cells (rag1, cd4, cd8), B cells (cd79a, igm) and NK cells (nccrp1, nkld) [46]. This revealed that both jak3+/ca and jak3ca/ca larvae displayed increased expression of T cell markers, reaching significance for rag1 and cd8 in both and cd4 in jak3ca/ca larvae, and NK cell markers, reaching significance for nccrp1 in jak3+/ca and nkld in jak3ca/ca larvae, whereas markers of HSCs and B cells were not significantly altered (Fig. 3A). Both jak3+/ca and jak3ca/ca larvae showed normal T cell and B cell rearrangement (Fig. 3B).
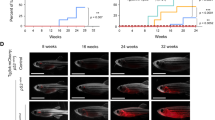
Effect of Jak3 A573V mutation on later hematopoiesis. A, B Gene expression analysis. Total RNA extracted from 28 dpf homozygous wild-type (jak3+/+), heterozygote (jak3+/ca) and homozygote (jak3ca/ca) mutant zebrafish was subjected to qRT2PCR analysis (A) with gene markers of hematopoietic stem cells (cmyb), T cells (rag1, cd4, cd8), B cells (cd79a, igm), and NK cells (nccrp1, nkld). Data are represented as relative fold change compared to homozygous wild-type (+/+) fish, with mean and SEM shown in red and statistical significance compared for Cq values normalized to control actb indicated (*p < 0.05; n = 3), or to RT-PCR (B) with primers specific for T cell receptor (TCR) β-chain (v(d)jcβ vb1.5, vb12, vb14.5) and B cell Ig heavy chain (igVH1, igVH4) rearrangements and actb as the control and analyzed by agarose gel electrophoresis. RT-negative controls yielded no products (data not shown) (n = 2). C–J Histological analysis. Giemsa stained blood cells (C–E) and kidney cells (G–I) of adult wild-type (jak3+/+) (C, G), heterozygote (jak3+/ca) (D, H) and homozygote (jak3ca/ca) (E, I) fish, with differential counts for blood (F) and kidney (J) performed (e: erythrocyte, eo: eosinophil, l: lymphocyte, n: neutrophil, p: precursor, m: macrophage) (n = 4–8; 100 × magnification; scale bar = 10 μm). K–P FACS analysis. Kidney cells from adult wild-type (jak3+/+) (K), heterozygote (jak3+/ca) (L) and homozygote (jak3ca/ca) (M) were subjected to FACS analysis, with lymphoid (N) myeloid (O) and precursor (P) populations quantified, with mean and SEM shown in red and level of statistical significance indicated (*p < 0.05, **p < 0.01; n = 5). Q Survival analysis. Wild-type (jak3+/+), heterozygote (jak3+/ca) and homozygote (jak3ca/ca) zebrafish were assessed weekly for survival, which is displayed as a Kaplan–Meier plot with level of statistical significance indicated (***p < 0.001, n = 35). R Gene expression analysis. Total RNA extracted from adult homozygous wild-type (jak3+/+), heterozygote (jak3+/ca) and homozygote (jak3ca/ca) zebrafish was subjected to qRT2PCR analysis with gene markers of hematopoietic stem cells (cmyb), T cells (tcra), B cells (cd79a), and NK cells (nccrp1), as described for panel A
Adult jak3+/+, jak3+/ca and jak3ca/ca zebrafish were also investigated. Cytological analysis of blood (Fig. 3C–E) and its quantitation by differential counting (Fig. 3F) revealed a significant increase in circulating lymphocyte numbers in jak3+/ca and jak3ca/ca in comparison to jak3+/+ fish. Analysis of the adult kidney, which serves a similar function to mammalian bone marrow with respect to hematopoiesis [7], identified a significant increase in lymphocyte numbers in both jak3+/ca and jak3ca/ca in comparison to jak3+/+ fish (Fig. 3G-J). Kidney cells were also subjected to flow cytometry analysis, with the lymphocyte (Fig. 3K-M, N) and precursor (Fig. 3K-M, P) populations both increased significantly in jak3+/ca and jak3ca/ca in comparison to jak3+/+ fish. In contrast, the myeloid (Fig. 3K-M, O) populations showed no statistically significant alterations. Expression analysis confirmed increased tcra+ T cells and nccrp1+ NK cells in this tissue in male fish (Fig. 3R).
The survival of jak3+/+, jak3+/ca and jak3ca/ca fish was also monitored. Mortality of jak3ca/ca fish was increased during the first 5 wpf compared to jak3+/+ and jak3+/ca fish (Fig. 3Q), but this stabilized such that these fish reached adulthood and were healthy and fecund (data not shown).
Molecular analysis of Jak3 A573V mutants
Other research has implicated various cytokine receptor signaling components in mediating the effects of JAK3 and mutants thereof [17, 20, 41]. The possession of a number of relevant zebrafish mutants made it possible to analyze these interactions genetically.
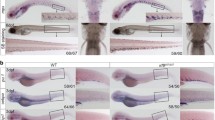
JAK3 is exclusively associated with the common cytokine receptor chain IL-2Rγc [18]. Zebrafish possesses two paralogues for the human IL2RG gene, with il2rga shown to have a conserved role in lymphopoiesis acting upstream of jak3 [59, 60]. Therefore, to elucidate the role of IL-2Rγc in mediating the effects of Jak3 A573V, fish carrying jak3ca alleles were crossed with those carrying knockout alleles for il2rga [59] to produce il2rga+/− jak3+/ca fish that were in-crossed to generate progeny that were assessed for rag1 expression and genotyped in parallel (Supp. Figure 1). As expected, for those embryos on an il2rga+/+ background, rag1 expression was significantly greater in jak3+/ca and jak3ca/ca compared to jak3+/+ embryos (Fig. 4A). For those on an il2rga−/− background, relative rag1 expression was significantly decreased for all jak3 genotypes, but importantly, there was no longer a significant difference between jak3+/+ and either jak3+/ca or jak3ca/ca embryos. This suggests an essential requirement for IL-2Rγc in mediating the effects of JAK3 A573V in vivo.
Molecular analysis of Jak3 A573V mutants. A-D. Genetic studies. Relative rag1 expression of jak3+/+, jak3+/ca and jak3ca/ca embryos on the indicated il2rga (A), jak1 (B), stat5.1 (C) and stat5.2 (D) genetic backgrounds, as detailed in Supp. Figure 1, 2, 3 and 4, respectively. Results for individual embryos of the relevant genotypes are shown along with mean and SEM in red and statistically significant differences indicated (*p < 0.05, **p < 0.01, ***p < 0.001, ns: not significant, n = 400). For simplicity, the results for some genotypes in panels A, B have been omitted. E Pharmacological studies. Relative rag1 expression of jak3+/+, jak3+/ca and jak3ca/ca embryos in the presence of the indicated Tofacitib concentrations. Results for individual embryos are shown along with mean and SEM in red and statistically significant differences indicated (***p < 0.001, ns: not significant, n = 400)
Amongst IL-2R family members, the ligand-specific receptor chains are associated with JAK1, which has been shown to be essential for signaling [20]. Zebrafish possesses a single jak1 orthologue [37], which has been demonstrated to play a conserved role in early lymphopoiesis [23]. Therefore, to evaluate the role of JAK1 in JAK3 A573V-mediated lymphoproliferation, jak3ca-carrying fish were crossed with jak1 mutant fish to produce jak1+/− jak3+/ca fish that were in-crossed to generate progeny for analysis (Supp. Figure 2). For those on a jak1+/+ background, relative rag1 expression was greater in jak3+/ca and jak3ca/ca embryos compared to jak3+/+ controls to a statistically significant extent (Fig. 4B). However, for those on a jak1−/− background, rag1 expression was significantly decreased for all jak3 genotypes and notably there was no significant difference observed between jak3+/+ and either jak3+/ca or jak3ca/ca embryos, confirming that JAK1 is also required for JAK3 A573V to exert its in vivo impacts.
STAT5 is strongly activated by JAK3 following stimulation of IL-2Rγc-containing receptors [34, 43], and is also activated by JAK3 mutants [17, 41]. Zebrafish possesses two STAT5 proteins encoded by stat5.1 and stat5.2 [33], both of which have been implicated downstream of il2rga/jak3 [60]. Since no differences between jak3+/ca and jak3ca/ca embryos had thus far been seen, a simpler crossing strategy was used, such that relative rag1 expression was compared between jak3+/+ and jak3+/ca genotypes on either a stat5.1 or stat5.2 homozygote mutant background compared to respective heterozygotes (Supp. Figures 3 and 4). As was the case on a wild-type background, a significant increase in rag1 was observed in jak3+/ca compared to jak3+/+ embryos on a heterozygote stat5.1+/− background embryos (Fig. 4C). However, no statistical difference in rag1 expression was observed in jak3+/ca compared to jak3+/+ embryos on a stat5.1−/− background. In contrast, a significant increase in rag1 was observed in jak3+/ca compared to jak3+/+ embryos on both stat5.2+/− and stat5.2−/− backgrounds (Fig. 4D). This suggests a differential requirement for STAT5 paralogues in mediating the effects of JAK3 A573V.
Several JAK inhibitors are under investigation for the treatment of hematopoietic diseases, including myeloproliferative neoplasms and autoimmune diseases, including the JAK3-specific Tofacitinib for treatment of rheumatoid arthritis [8, 27]. The efficacy of Tofacitinib was tested on jak3+/ca in comparison to jak3+/+ embryos with treatment from 3 to 5 dpf followed by analysis of relative rag1 staining (Fig. 4E). A significant decrease in rag1 expression was seen in jak3+/+ embryos treated with 30 nM and 60 nM Tofacitinib compared to the untreated group, with no significant difference between these concentrations, as described previously [60]. The extent of rag1 staining was also reduced in jak3+/ca following Tofacitinib treatment, although there was an even greater decrease with 60 nM compared to 30 nM. Collectively, these data show that the Jak3 A573V mutant was susceptible to Tofacitinib, although may require a higher dose for maximal inhibition.
Discussion
Lymphoid development relies heavily on the IL-2R family of cytokine receptors, which includes IL-2R, IL-4R, IL-7R, IL-9R, IL-15R and IL-21R [31]. Each of these share the common IL-2Rγc that signals via JAK3, as well as a ligand-specific chain that signals through JAK1 and sometimes a third receptor chain, with STAT5 a key downstream effector [30, 36]. Zebrafish has been employed extensively to study blood and immune cell development and its disruption in malignancy [46, 51, 55]. Importantly, it possesses many of the constituent chains of the IL-2R family, including a duplicated IL-2Rγc [38], as well as homologues of JAK1, JAK3 and STAT5 proteins [33, 37], with conserved roles described for IL-2Rγc, JAK1, JAK3 and STAT5 in embryonic lymphopoiesis [23, 60]. Constitutively activating mutations in JAK3 (or JAK1) are associated with a range of hematological malignancies [3, 6, 19, 44, 48], with the PKD mutations A572V and A573V common in several forms of disease [3, 6, 27, 49, 64]. Since the sequence around A572/A573 was highly conserved, these specific mutations could be readily recapitulated in the zebrafish protein. The zebrafish Jak3 A573V mutant elicited greater activation of zebrafish Stat5.1 in cell culture than the A572V mutant and was also able to significantly increase T cells when introduced into zebrafish embryos. Therefore, to study the impact of enhanced Jak3 activation in a physiologically-relevant context across the life course, a zebrafish Jak3 A573V mutant line was generated using CRISPR–Cas9. This mutant was broadly characterized during embryonic, larval and adult hematopoiesis, with its genetic interaction with other IL-2R family signaling components and sensitivity to a JAK inhibitor analyzed.
The impacts of the Jak3 A573V mutation were evident already in early lymphoid precursors in the developing embryo through elevated ikzf1 staining at 3.5 dpf. This continued at 5 dpf, when the more mature T cell markers rag1, tcra and lck were also significantly elevated, with analysis of blood smears confirming increased numbers of circulating lymphocytes. Elevated T and NK cell markers were evident in juvenile fish at 28 dpf, but no significant effects on B cells were observed. Adult Jak3 A573V mutants also possessed increased lymphocytes in the blood and kidney, with elevated T and NK cells confirmed in adult male kidney. It has been suggested that the presence of wild-type JAK3 suppresses the effects of mutant JAK3s, with patient JAK3 mutations often being homozygous or compound heterozygous [16, 40]. However, zebrafish carrying two copies of the allele encoding Jak3 A573V mutant did not show more severe impacts on lymphopoiesis than those with one copy, although reduced survival of homozygote mutants compared to wild-type fish was observed that was not seen with heterozygotes. This could reflect a tendency to progress to a more severe disorder, although no evidence of overt leukemia has been identified. Together these data are consistent with constitutively active JAK3 eliciting a mild lymphoproliferative disorder rather than overt leukemia, with mutation of a single allele sufficient to achieve this.
Mouse transplantation models have been widely used to study JAK3 mutants. For example, retroviral transduction of mouse bone marrow cells to overexpress human JAK3 A572V followed by transplantation into lethally irradiated mice resulted in megakaryocytic hyperplasia and lymphoproliferative disorders [11], including a T-ALL like disease, with ligand-independent proliferation of T cells [17]. However, these studies involved overexpression of the mutant JAK3 across multiple cell lineages also expressing wild-type mouse Jak3, in contrast to introducing the mutation into the endogenous protein expressed from its native promoter within its normal cellular context. Notably, a mouse Jak3 A572V knock-in model exhibited a more similar phenotype to the zebrafish model, with progressive expansion of CD8+ T cells and minor skin pathology, with a fully penetrant, lethal disease only manifest in cooperation with partial trisomy 21 [57]. These more physiologically relevant models emphasize the importance of additional co-operating mutations for leukemic transformation, as is observed in humans [3, 6, 17]. Key co-operating mutations include those in genes encoding the epigenetic regulators SUZ12 [9] and PHF6 [69] as well as the transcription factors RUNX1 [2] and HOXA9 [15], with the latter co-occupying similar genomic loci as STAT5. One explanation for the absence of significant B cell involvement in either model is that while JAK3 is a ‘driver’ mutation in T cell malignancy, it is considered more of a ‘co-operating’ gene in B cell malignancy, as described in the context of PAX5 fusions [24] and SPI1 deletions [5]. Crossing the zebrafish Jak3 A573V mutant with lines harboring other oncogenic mutations would yield definitive insights.
Previous research has demonstrated that most JAK3 mutants require IL-2Rγc to activate downstream signaling and factor-independent growth [1], and both IL-2Rγc and JAK1 for maximal activation of downstream signaling [17, 41, 54]. The kinase activity of JAK1 has been shown to be essential to facilitate this [20, 41, 54], with JAK1 constitutively activated in the presence of JAK3 mutants [16, 41]. In this study, genetic analysis has revealed that the requirement for IL-2Rγc and JAK1 extends to the in vivo environment, with both found to be essential for mutant JAK3 to mediate the expansion of embryonic T lymphocytes. Such a role for IL-2Rγc has also been observed in the mouse Jak3 A572V knock-in model [57]. These data are consistent with JAK3 mutations occurring in concert with JAK1 mutations [3, 6, 19, 49], with JAK1 and JAK3 mutations able to synergize in vitro [20].
STAT5 proteins are constitutively activated by JAK3 mutants [16, 41]. The data presented here demonstrates for the first time a requirement for downstream STAT5 in mediating the effects of JAK3 mutants—and specifically the zebrafish Stat5.1 paralogue, which has been identified as the functional equivalent of STAT5B rather than STAT5A [68]. These results are compatible with studies showing STAT5B exerting a greater effect on cytokine-driven lymphocyte proliferation than STAT5A [22, 47]. Moreover, activating STAT5B mutations have been identified in a variety of hematological neoplasms, being identified as likely driver mutations in various T cell neoplasms [14, 52, 63], with JAK3 and STAT5B in the same mutation cluster in ALL [40]. Interestingly STAT5B but not STAT5A has also been demonstrated to facilitate BCR/ABL-induced leukemic transformation [26], and we have previously shown that a zebrafish Stat5.1 N649H mutant can reproduce features of STAT5B N649H-mediated disease that impacts both lymphoid and myeloid lineages [32]. Collectively, this highlights a specific, and evolutionarily conserved role for STAT5B/Stat5.1 in hematological malignancy.
The identification of JAK mutations in a broad range of hematological malignancies and proliferative disorders has promoted the development of a myriad of pharmacological inhibitors [42, 53]. In this study, we show that Jak3 A573V-mediated lymphocyte expansion was sensitive to Tofacitib, a pan-JAK inhibitor shown to be efficacious in human clinical trials [12]—confirming similar results in the mouse Jak3 A572V model [57] and a variety of in vitro studies [8, 16, 27]. In fact, one of these studies identified STAT5 activation as showing the highest sensitivity to the effects of this inhibitor [16], further emphasizing its key role. Together this not only serves to confirm the applicability of this model to pharmacological testing, but also indicates that approaches targeting STAT5B would also be appropriate to consider.
Conclusion
This study demonstrated that a single JAK3 A573V allele expressed from its endogenous locus was able to enhance zebrafish lymphopoiesis throughout the life-course, producing a sustained increase in T cells, as well as affecting NK cells, but not B cells. This was mediated via an IL-2Rγc/JAK1/JAK3/STAT5B signaling pathway and was sensitive to Tofacitinib. This enhances our understanding of oncogenic JAK3 mutations in vivo and creates a novel model to underpin further translational investigations.
Availability of data and materials
All data generated or analyzed during this study are included in this published article (and its supplementary information files).
Abbreviations
- AMKL:
-
Acute megakaryoblastic leukemia
- ca:
-
Constitutively activating
- Cas9:
-
CRISPR-associated 9
- CRISPR:
-
Clustered regularly interspaced short palindromic repeats
- CTCL:
-
Cutaneous T cell leukemia
- dpf:
-
Days post-fertilization
- FSC:
-
Forward scatter
- HDR:
-
Homology-directed repair
- HRM:
-
High-resolution melt
- IL:
-
Interleukin
- IL-2Rγc:
-
IL-2 receptor gamma common
- JAK:
-
Janus kinase
- NKTCL:
-
Natural killer T cell lymphoma
- PKD:
-
Pseudokinase domain
- qRT2-PCR:
-
Quantitative real-time RT-PCR
- RFLP:
-
Restriction fragment length polymorphism
- RT-PCR:
-
Reverse transcription-polymerase chain reaction
- SEM:
-
Standard error of the mean
- SSC:
-
Side scatter
- STAT:
-
Signal transducer and activator of transcription
- T-ALL:
-
T cell acute lymphoblastic leukemia
- T-PLL:
-
T cell prolymphocytic leukemia
- WISH:
-
Whole-mount in situ hybridization
References
Agarwal A, MacKenzie RJ, Eide CA, Davare MA, Watanabe-Smith K, Tognon CE et al (2015) Functional RNAi screen targeting cytokine and growth factor receptors reveals oncorequisite role for interleukin-2 gamma receptor in JAK3-mutation-positive leukemia. Oncogene 34:2991–2999
Avagyan S (2021) Brown AL To T or not to B: germline RUNX1 mutation preferences in pediatric ALL predisposition. J Clin Investig 131:e152464
Bains T, Heinrich M, Loriaux M, Beadling C, Nelson D, Warrick A et al (2012) Newly described activating JAK3 mutations in T-cell acute lymphoblastic leukemia. Leukemia 26:2144–2146
Basheer F, Rasighaemi P, Liongue C (2019) Ward AC Zebrafish granulocyte colony-stimulating factor receptor maintains neutrophil number and function throughout the life span. Infect Immun 87:e00793-e818
Batista CR, Lim M, Laramee AS, Abu-Sardanah F, Xu LS, Hossain R et al (2018) Driver mutations in Janus kinases in a mouse model of B-cell leukemia induced by deletion of PU.1 and Spi-B. Blood Adv 2:2798–2810
Bellanger D, Jacquemin V, Chopin M, Pierron G, Bernard O, Ghysdael J et al (2014) Recurrent JAK1 and JAK3 somatic mutations in T-cell prolymphocytic leukemia. Leukemia 28:417–419
Bertrand JY, Kim AD, Teng S (2008) Traver D CD41+ cmyb+ precursors colonize the zebrafish pronephros by a novel migration route to initiate adult hematopoiesis. Development 135:1853–1862
Bouchekioua A, Scourzic L, De Wever O, Zhang Y, Cervera P, Aline-Fardin A et al (2014) JAK3 deregulation by activating mutations confers invasive growth advantage in extranodal nasal-type natural killer cell lymphoma. Leukemia 28:338–348
Broux M, Prieto C, Demeyer S, Vanden Bempt M, Alberti-Servera L, Lodewijckx I et al (2019) Suz12 inactivation cooperates with JAK3 mutant signaling in the development of T-cell acute lymphoblastic leukemia. Blood 134:1323–1336
Candotti F, O’Shea JJ (1998) Villa A Severe combined immune deficiencies due to defects of the common gamma chain-JAK3 signaling pathway. Springer Semin Immunopathol 19:401–415
Cornejo MG, Kharas MG, Werneck MB, Le Bras S, Moore SA, Ball B et al (2009) Constitutive JAK3 activation induces lymphoproliferative syndromes in murine bone marrow transplantation models. Blood 113:2746–2754
Damsky W, Peterson D, Ramseier J, Al-Bawardy B, Chun H, Proctor D et al (2021) The emerging role of Janus kinase inhibitors in the treatment of autoimmune and inflammatory diseases. J Allergy Clin Immunol 147:814–826
Danilova N, Hohman VS, Sacher F, Ota T, Willett CE (2004) Steiner LA T cells and the thymus in developing zebrafish. Dev Comp Immunol 28:755–767
De Bie J, Demeyer S, Alberti-Servera L, Geerdens E, Segers H, Broux M et al (2018) Single-cell sequencing reveals the origin and the order of mutation acquisition in T-cell acute lymphoblastic leukemia. Leukemia 32:1358–1369
de Bock CE, Demeyer S, Degryse S, Verbeke D, Sweron B, Gielen O et al (2018) HOXA9 cooperates with activated JAK/STAT signaling to drive leukemia development. Cancer Discov 8:616–631
Degryse S, Bornschein S, de Bock CE, Leroy E, Vanden Bempt M, Demeyer S et al (2018) Mutant JAK3 signaling is increased by loss of wild-type JAK3 or by acquisition of secondary JAK3 mutations in T-ALL. Blood 131:421–425
Degryse S, De Bock CE, Cox L, Demeyer S, Gielen O, Mentens N et al (2014) JAK3 mutants transform hematopoietic cells through JAK1 activation, causing T-cell acute lymphoblastic leukemia in a mouse model. Blood 124:3092–3100
Ghoreschi K, Laurence A, O’Shea JJ (2009) Janus kinases in immune cell signaling. Immunol Rev 228:273–287
Greenplate A, Wang K, Tripathi RM, Palma N, Ali SM, Stephens PJ et al (2018) Genomic profiling of T-cell neoplasms reveals frequent JAK1 and JAK3 mutations with clonal evasion from targeted therapies. JCO Precis Oncol 2018:PO.17.00019
Haan C, Rolvering C, Raulf F, Kapp M, Druckes P, Thoma G et al (2011) Jak1 has a dominant role over Jak3 in signal transduction through gammac-containing cytokine receptors. Chem Biol 18:314–323
Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD et al (2013) Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 31:227–229
Imada K, Bloom ET, Nakajima H, Horvath-Arcidiacono JA, Udy GB, Davey HW et al (1998) Stat5b is essential for natural killer cell-mediated proliferation and cytolytic activity. J Exp Med 188:2067–2074
Iwanami N, Mateos F, Hess I, Riffel N, Soza-Ried C, Schorpp M et al (2011) Genetic evidence for an evolutionarily conserved role of IL-7 signaling in T cell development of zebrafish. J Immunol 186:7060–7066
Jamrog L, Chemin G, Fregona V, Coster L, Pasquet M, Oudinet C et al (2018) PAX5-ELN oncoprotein promotes multistep B-cell acute lymphoblastic leukemia in mice. Proc Natl Acad Sci USA 115:10357–10362
Kiyoi H, Yamaji S, Kojima S, Naoe T (2007) JAK3 mutations occur in acute megakaryoblastic leukemia both in Down syndrome children and non-Down syndrome adults. Leukemia 21:574–576
Kollmann S, Grundschober E, Maurer B, Warsch W, Grausenburger R, Edlinger L et al (2019) Twins with different personalities: STAT5B-but not STAT5A-has a key role in BCR/ABL-induced leukemia. Leukemia 33:1583–1597
Koo GC, Tan SY, Tang T, Poon SL, Allen GE, Tan L et al (2012) Janus kinase 3-activating mutations identified in natural killer/T-cell lymphoma. Cancer Disc 2:591–597
Langenau DM, Ferrando AA, Traver D, Kutok JL, Hezel JP, Kanki JP et al (2004) In vivo tracking of T cell development, ablation, and engraftment in transgenic zebrafish. Proc Natl Acad Sci USA 101:7369–7374
Langenau DM, Zon LI (2005) The zebrafish: a new model of T-cell and thymic development. Nat Rev Immunol 5:307–317
Leonard WJ, Lin J-X (2000) Cytokine receptor signaling pathways. J Allergy Clin Immunol 105:877–888
Leonard WJ, Lin JX, O’Shea JJ (2019) The gammac family of cytokines: basic biology to therapeutic ramifications. Immunity 50:832–850
Lewis RS, Stephenson SEM, Ward AC (2006) Constitutive activation of zebrafish stat5 expands hematopoietic cell populations in vivo. Exp Hematol 34:179–187
Lewis RS, Ward AC (2004) Conservation, duplication and divergence of the zebrafish stat5 genes. Gene 338:65–74
Liao W, Lin JX, Leonard WJ (2013) Interleukin-2 at the crossroads of effector responses, tolerance, and immunotherapy. Immunity 38:13–25
Lieschke GJ, Oates AC, Crowhurst MO, Ward AC, Layton JE (2001) Morphologic and functional characterization of granulocytes and macrophages in embryonic and adult zebrafish. Blood 98:3087–3096
Lin JX, Leonard WJ (2000) The role of Stat5a and Stat5b in signaling by IL-2 family cytokines. Oncogene 19:2566–2576
Liongue C, O’Sullivan LA, Trengove MC (2012) Ward AC Evolution of JAK-STAT pathway components: mechanisms and role in immune system development. PLoS ONE 7:e32777
Liongue C, Ward AC (2007) Evolution of class I cytokine receptors. BMC Evol Biol 7:120
Liu F, Wen Z (2002) Cloning and expression pattern of the lysozyme C gene in zebrafish. Mech Dev 113:69–72
Liu Y, Easton J, Shao Y, Maciaszek J, Wang Z, Wilkinson MR et al (2017) The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet 49:1211–1218
Losdyck E, Hornakova T, Springuel L, Degryse S, Gielen O, Cools J et al (2015) Distinct acute lymphoblastic leukemia (ALL)-associated Janus kinase 3 (JAK3) mutants exhibit different cytokine-receptor requirements and JAK inhibitor specificities. J Biol Chem 290:29022–29034
Luo Y, Alexander M, Gadina M, O’Shea JJ, Meylan F (2021) Schwartz DM JAK-STAT signaling in human disease: from genetic syndromes to clinical inhibition. J Allergy Clin Immunol 148:911–925
Malek TR, Castro I (2010) Interleukin-2 receptor signaling: at the interface between tolerance and immunity. Immunity 33:153–165
Malinge S, Ragu C, Della-Valle V, Pisani D, Constantinescu SN, Perez C et al (2008) Activating mutations in human acute megakaryoblastic leukemia. Blood 112:4220–4226
McGirt LY, Jia P, Baerenwald DA, Duszynski RJ, Dahlman KB, Zic JA et al (2015) Whole-genome sequencing reveals oncogenic mutations in mycosis fungoides. Blood 126:508–519
Moore FE, Garcia EG, Lobbardi R, Jain E, Tang Q, Moore JC et al (2016) Single-cell transcriptional analysis of normal, aberrant, and malignant hematopoiesis in zebrafish. J Exp Med 213:979–992
Moriggl R, Topham DJ, Teglund S, Sexl V, McKay C, Wang D et al (1999) Stat5 is required for IL-2-induced cell cycle progression of peripheral T cells. Immunity 10:249–259
Mullighan CG, Zhang J, Harvey RC, Collins-Underwood JR, Schulman BA, Phillips LA et al (2009) JAK mutations in high-risk childhood acute lymphoblastic leukemia. Proc Natl Acad Sci USA 106:9414–9418
Nicolae A, Xi L, Pham TH, Pham TA, Navarro W, Meeker HG et al (2016) Mutations in the JAK/STAT and RAS signaling pathways are common in intestinal T-cell lymphomas. Leukemia 30:2245–2247
O’Sullivan LA, Liongue C, Lewis RS, Stephenson SEM, Ward AC (2007) Cytokine receptor signaling through the Jak/Stat/Socs pathway in disease. Mol Immunol 44:2497–2506
Page DM, Wittamer V, Bertrand JY, Lewis KL, Pratt DN, Delgado N et al (2013) An evolutionarily conserved program of B-cell development and activation in zebrafish. Blood 122:e1–e11
Pham HTT, Maurer B, Prchal-Murphy M, Grausenburger R, Grundschober E, Javaheri T et al (2018) STAT5BN642H is a driver mutation for T cell neoplasia. J Clin Investig 128:387–401
Raivola J, Haikarainen T, Abraham BG, Silvennoinen O (2021) Janus kinases in leukemia. Cancers (Basel) 13:800
Raivola J, Hammaren HM, Virtanen AT, Bulleeraz V, Ward AC, Silvennoinen O (2018) Hyperactivation of oncogenic JAK3 mutants depend on ATP binding to the pseudokinase domain. Front Oncol 8:560
Rasighaemi P, Basheer F, Liongue C, Liongue C, Ward AC (2015) Zebrafish as a model for leukemia and other hematopoietic disorders. J Hematol Oncol 8:29
Riera L, Lasorsa E, Bonello L, Sismondi F, Tondat F, Di Bello C et al (2011) Description of a novel Janus kinase 3 P132A mutation in acute megakaryoblastic leukemia and demonstration of previously reported Janus kinase 3 mutations in normal subjects. Leuk Lymphoma 52:1742–1750
Rivera-Munoz P, Laurent AP, Siret A, Lopez CK, Ignacimouttou C, Cornejo MG et al (2018) Partial trisomy 21 contributes to T-cell malignancies induced by JAK3-activating mutations in murine models. Blood Adv 2:1616–1627
Robb L (2007) Cytokine receptors and hematopoietic differentiation. Oncogene 26:6715–6723
Sertori R, Jones R, Basheer F, Rivera L, Dawson S, Loke S et al (2022) Generation and characterization of a zebrafish IL-2Rgc SCID model. Int J Mol Sci 23:2385
Sertori R, Liongue C, Basheer F, Lewis KL, Rasighaemi P, de Coninck D et al (2016) Conserved IL-2Rgc signaling mediates lymphopoiesis in zebrafish. J Immunol 196:135–143
Sertori R, Trengove M, Basheer F, Ward AC, Liongue C (2016) Genome editing in zebrafish: a practical overview. Brief Funct Genom 15:322–330
Thisse C, Thisse B (2008) High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc 3:59–69
Wahnschaffe L, Braun T, Timonen S, Giri AK, Schrader A, Wagle P et al (2019) JAK/STAT-activating genomic alterations are a hallmark of T-PLL. Cancers (Basel) 11:1833
Walters DK, Mercher T, Gu TL, O’Hare T, Tyner JW, Loriaux M et al (2006) Activating alleles of JAK3 in acute megakaryoblastic leukemia. Cancer Cell 10:65–75
Westerfield M (1995) The zebrafish book, 3rd edn. University of Oregon Press, Eugene
Willett CE, Kawasaki H, Amemiya CT, Lin S, Steiner LA (2001) Ikaros expression as a marker for lymphoid progenitors during zebrafish development. Dev Dyn 222:694–698
Willett CE, Zapata A, Hopkins N, Steiner LA (1997) Expression of zebrafish rag genes during early development identifies the thymus. Dev Biol 182:331–341
Xiong S, Mei J, Huang P, Jing J, Li Z, Kang J et al (2017) Essential roles of stat5.1/stat5b in controlling fish somatic growth. J Genet Genom 44:577–585
Yuan S, Wang X, Hou S, Guo T, Lan Y, Yang S et al (2022) PHF6 and JAK3 mutations cooperate to drive T-cell acute lymphoblastic leukemia progression. Leukemia 36:370–382
Acknowledgements
The authors would like to thank the Deakin University Animal House staff for superb aquarium management, and Somayyeh Heidary, Nagendra Awasthi, Mohammed Luban Sobah and Parisa Rasighaemi for technical assistance.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions. The authors recognize the support of an International Research Scholarship (FB) and project costs from Deakin University. The funder had no role in study design, data collection and interpretation or manuscript preparation.
Author information
Authors and Affiliations
Contributions
ACW conceived the project. FB generated the zebrafish JAK3 mutants and performed most of its in vivo and in vitro characterization, VB generated and analyzed the various double mutants, and VQTN performed the inhibitor studies and confirmed some other results. ACW and CL co-supervised FB, VB and VQTN. All authors contributed to data interpretation and drafting of the paper, and all approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethics approval and consent to participate
This study was approved by the Deakin University Animal Ethics Committee.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
18_2022_4361_MOESM1_ESM.jpg
Supplementary Figure 1: Investigating the role of IL-2Rγc in mediating the effects of Jak3 A573V. A. Embryos were raised from il2rga+/− jak3+/ca × il2rga+/− jak3+/ca crosses until 5 dpf then subjected to WISH with rag1. Individual embryos were imaged to determine the area of rag1 expression and then genomic DNA was extracted for PCR-based genotyping, using HRM analysis for jak3 (lower left) and RFLP analysis for il2rga (lower right) using a PCR with il2rga-specific primers (5’- CGAAGACTGTCCTGAATATGAGAC, 5’- TCTGGTCAGTCCTGTAACGAAC) followed by NdeI digestion. B-G. Images of rag1 staining for representative il2rga+/+ jak3+/+ (B), il2rga−/− jak3+/+ (C), il2rga+/+ jak3+/ca (D), il2rga−/− jak3+/ca (E), il2rga+/+ jak3ca/ca (F) and il2rga−/− jak3ca/ca (G) embryos with rag1 expression indicated by arrows (JPG 7059 kb)
18_2022_4361_MOESM2_ESM.jpg
Supplementary Figure 2: Investigating the role of Jak1 in mediating the effects of Jak3 A573V. A. Embryos were raised from jak1+/− jak3+/ca × jak1+/− jak3+/ca crosses until 5 dpf then subjected to WISH with rag1. Individual embryos were imaged to determine the area of rag1 expression and then genomic DNA was extracted for PCR-based genotyping, using HRM analysis for jak3 (lower left) and jak1 (lower right) with jak1-specific primers (5’-CAGGCATTCTTTGAGACCGC, 5’-GGGTACTTACTCTCCTGGTGACG). B-G. Images of rag1 staining for representative jak1+/+ jak3+/+ (B), jak1−/− jak3+/+ (C), jak1+/+ jak3+/ca (D), jak1−/− jak3+/ca (E), jak1+/+ jak3ca/ca (F) and jak1−/− jak3ca/ca (G) embryos with rag1 expression indicated by arrows (JPG 7856 kb)
18_2022_4361_MOESM3_ESM.jpg
Supplementary Figure 3: Investigating the role of Stat5.1 in mediating the effects of Jak3 A573V. A. Embryos were raised from a stat5.1+/− jak3+/ca × stat5.1−/− jak3+/+ cross until 5 dpf then subjected to WISH with rag1.Embryos were imaged to determine the area of rag1 expression and then genomic DNA was extracted for PCR-based genotyping, using HRM analysis for jak3 (lower left) and PCR for stat5.1 (lower right) with stat5.1-specific primers (5’-GTGGGCGGGTTAATGGACAG, 5’-TACACGCATACCCTGTATTCTGAG). B-E: Images of rag1 expression for representative stat5.1+/− jak3+/+ (B), stat5.1+/− jak3+/ca (C), stat5.1−/− jak3+/+ (D), and stat5.1−/− jak3+/ca (E) embryos with rag1 expression indicated by arrows (JPG 4091 kb)
18_2022_4361_MOESM4_ESM.jpg
Supplementary Figure 4: Investigating the role of Stat5.2 in mediating the effects of Jak3 A573V. A. Embryos were raised from a stat5.2+/− jak3+/ca × stat5.2−/− jak3+/+ cross until 5 dpf then subjected to WISH with rag1.Embryos were imaged to determine the area of rag1 expression and then genomic DNA was extracted for PCR-based genotyping, using HRM analysis for jak3 (lower left) and PCR for stat5.2 (lower right) with stat5.2-specific primers (5’- CAGCAGTCCAGGTTCAGGTC, 5’-GATCATACCCTGTATCCTCAAACTC). B-E: Images of rag1 expression for representative stat5.2+/− jak3+/+ (B), stat5.2+/− jak3+/ca (C), stat5.2−/− jak3+/+ (D), and stat5.2−/− jak3+/ca (E) embryos with rag1 expression indicated by arrows (JPG 5797 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Basheer, F., Bulleeraz, V., Ngo, V.Q.T. et al. In vivo impact of JAK3 A573V mutation revealed using zebrafish. Cell. Mol. Life Sci. 79, 322 (2022). https://doi.org/10.1007/s00018-022-04361-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-022-04361-8