Abstract
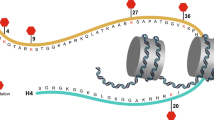
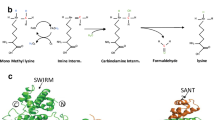
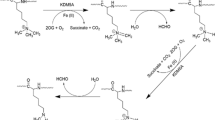
NSD1, NSD2, and NSD3 constitute the nuclear receptor-binding SET Domain (NSD) family of histone 3 lysine 36 (H3K36) methyltransferases. These structurally similar enzymes mono- and di-methylate H3K36, which contribute to the maintenance of chromatin integrity and regulate the expression of genes that control cell division, apoptosis, DNA repair, and epithelial–mesenchymal transition (EMT). Aberrant expression or mutation of members of the NSD family is associated with developmental defects and the occurrence of some types of cancer. In this review, we discuss the effect of alterations in NSDs on cancer patient’s prognosis and response to treatment. We summarize the current understanding of the biological functions of NSD proteins, focusing on their activities and the role in the formation and progression in solid tumors biology, as well as how it depends on tumor etiologies. This review also discusses ongoing efforts to develop NSD inhibitors as a promising new class of cancer therapeutic agents.





Similar content being viewed by others
Data availability
Not applicable.
References
Martin C, Zhang Y (2005) The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol 6(11):838–849
Strahl BD, Allis CD (2000) The language of covalent histone modifications. Nature 403(6765):41–45
Husmann D, Gozani O (2019) Histone lysine methyltransferases in biology and disease. Nat Struct Mol Biol 26(10):880–889
Zaghi M, Broccoli V, Sessa A (2019) H3K36 methylation in neural development and associated diseases. Front Genet 10:1291
Kim A, Kiefer CM, Dean A (2007) Distinctive signatures of histone methylation in transcribed coding and noncoding human beta-globin sequences. Mol Cell Biol 27(4):1271–1279
Bannister AJ et al (2005) Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem 280(18):17732–17736
Fang D et al (2016) The histone H3.3K36M mutation reprograms the epigenome of chondroblastomas. Science 352(6291):1344–1348
Li J, Ahn JH, Wang GG (2019) Understanding histone H3 lysine 36 methylation and its deregulation in disease. Cell Mol Life Sci 76(15):2899–2916
Popovic R et al (2014) Histone methyltransferase MMSET/NSD2 alters EZH2 binding and reprograms the myeloma epigenome through global and focal changes in H3K36 and H3K27 methylation. PLoS Genet 10(9):e1004566
Rogawski DS, Grembecka J, Cierpicki T (2016) H3K36 methyltransferases as cancer drug targets: rationale and perspectives for inhibitor development. Future Med Chem 8(13):1589–1607
Tsukada Y-I et al (2006) Histone demethylation by a family of JmjC domain-containing proteins. Nature 439(7078):811–816
Hyun K et al (2017) Writing, erasing and reading histone lysine methylations. Exp Mol Med 49(4):e324–e324
Whetstine JR et al (2006) Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125(3):467–481
Wagner EJ, Carpenter PB (2012) Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol 13(2):115–126
Bennett RL et al (2017) The role of nuclear receptor-binding SET domain family histone lysine methyltransferases in cancer. Cold Spring Harb Perspect Med 7(6):a026708
Rayasam GV (2003) NSD1 is essential for early post-implantation development and has a catalytically active SET domain. EMBO J 22(12):3153–3163
Shirane K et al (2020) NSD1-deposited H3K36me2 directs de novo methylation in the mouse male germline and counteracts Polycomb-associated silencing. Nat Genet 52(10):1088–1098
Kurotaki N et al (2002) Haploinsufficiency of NSD1 causes Sotos syndrome. Nat Genet 30(4):365–366
Baujat G et al (2004) Paradoxical NSD1 mutations in Beckwith–Wiedemann syndrome and 11p15 anomalies in Sotos syndrome. Am J Hum Genet 74(4):715–720
Nimura K et al (2009) A histone H3 lysine 36 trimethyltransferase links Nkx2-5 to Wolf–Hirschhorn syndrome. Nature 460(7252):287–291
Campos-Sanchez E et al (2017) Wolf–Hirschhorn syndrome candidate 1 is necessary for correct hematopoietic and B cell development. Cell Rep 19(8):1586–1601
Stec I et al (1998) WHSC1, a 90 kb SET domain-containing gene, expressed in early development and homologous to a Drosophila dysmorphy gene maps in the Wolf–Hirschhorn syndrome critical region and is fused to IgH in t(4;14) multiple myeloma. Hum Mol Genet 7(7):1071–1082
Adams MS, Gammill LS, Bronner-Fraser M (2008) Discovery of transcription factors and other candidate regulators of neural crest development. Dev Dyn 237(4):1021–1033
Jacques-Fricke BT, Gammill LS (2014) Neural crest specification and migration independently require NSD3-related lysine methyltransferase activity. Mol Biol Cell 25(25):4174–4186
Jacques-Fricke BT et al (2021) Profiling NSD3-dependent neural crest gene expression reveals known and novel candidate regulatory factors. Dev Biol 475:118–130
Jaju RJ et al (2001) A novel gene, NSD1, is fused to NUP98 in the t(5;11)(q35;p15.5) in de novo childhood acute myeloid leukemia. Blood 98(4):1264–1267
Panarello C, Rosanda C, Morerio C (2002) Cryptic translocation t(5;11)(q35;p15.5) with involvement of the NSD1 and NUP98 genes without 5q deletion in childhood acute myeloid leukemia. Genes Chromosomes Cancer 35(3):277–281
Angrand PO et al (2001) NSD3, a new SET domain-containing gene, maps to 8p12 and is amplified in human breast cancer cell lines. Genomics 74(1):79–88
Saloura V et al (2018) The role of protein methyltransferases as potential novel therapeutic targets in squamous cell carcinoma of the head and neck. Oral Oncol 81:100–108
Lucio-Eterovic AK et al (2010) Role for the nuclear receptor-binding SET domain protein 1 (NSD1) methyltransferase in coordinating lysine 36 methylation at histone 3 with RNA polymerase II function. Proc Natl Acad Sci 107(39):16952–16957
He C et al (2013) The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition. J Biol Chem 288(7):4692–4703
Qiao Q et al (2011) The structure of NSD1 reveals an autoregulatory mechanism underlying histone H3K36 methylation. J Biol Chem 286(10):8361–8368
Vougiouklakis T et al (2015) The NSD family of protein methyltransferases in human cancer. Epigenomics 7(5):863–874
Garlisi CG et al (2001) A unique mRNA initiated within a middle intron of WHSC1/MMSET encodes a DNA binding protein that suppresses human IL-5 transcription. Am J Respir Cell Mol Biol 24(1):90–98
Woo Park J et al (2015) RE-IIBP methylates H3K79 and induces MEIS1-mediated apoptosis via H2BK120 ubiquitination by RNF20. Sci Rep 5:12485
Kim SM et al (2006) Characterization of a novel WHSC1-associated SET domain protein with H3K4 and H3K27 methyltransferase activity. Biochem Biophys Res Commun 345(1):318–323
Graham SE, Tweedy SE, Carlson HA (2016) Dynamic behavior of the post-SET loop region of NSD1: implications for histone binding and drug development. Protein Sci 25(5):1021–1029
Morishita M, Mevius D, di Luccio E (2014) In vitro histone lysine methylation by NSD1, NSD2/MMSET/WHSC1 and NSD3/WHSC1L. BMC Struct Biol 14:25
Li W et al (2020) Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases. Nature 590:498–503
Han D et al (2019) Lysine methylation of transcription factors in cancer. Cell Death Dis 10(4):290
Bannister AJ, Kouzarides T (2011) Regulation of chromatin by histone modifications. Cell Res 21(3):381–395
Schmitges FW et al (2011) Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell 42(3):330–341
Yuan W et al (2011) H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J Biol Chem 286(10):7983–7989
Farhangdoost N et al (2021) Chromatin dysregulation associated with NSD1 mutation in head and neck squamous cell carcinoma. Cell Rep 34(8):108769
Streubel G et al (2018) The H3K36me2 methyltransferase Nsd1 demarcates PRC2-mediated H3K27me2 and H3K27me3 domains in embryonic stem cells. Mol Cell 70(2):371-379.e5
Woo H et al (2017) Modulation of gene expression dynamics by co-transcriptional histone methylations. Exp Mol Med 49(4):e326–e326
Carrozza MJ et al (2005) Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123(4):581–592
Keogh MC et al (2005) Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123(4):593–605
Li B et al (2007) Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes Dev 21(11):1422–1430
Fang Y et al (2021) The H3K36me2 methyltransferase NSD1 modulates H3K27ac at active enhancers to safeguard gene expression. Nucleic Acids Res 49(11):6281–6295
Fnu S et al (2011) Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc Natl Acad Sci U S A 108(2):540–545
De Krijger I et al (2020) H3K36 dimethylation by MMSET promotes classical non-homologous end-joining at unprotected telomeres. Oncogene 39(25):4814–4827
Hajdu I et al (2011) Wolf–Hirschhorn syndrome candidate 1 is involved in the cellular response to DNA damage. Proc Natl Acad Sci U S A 108(32):13130–13134
Zhang J et al (2019) PTEN methylation by NSD2 controls cellular sensitivity to DNA damage. Cancer Discov 9(9):1306–1323
Mah LJ, El-Osta A, Karagiannis TC (2010) γH2AX: a sensitive molecular marker of DNA damage and repair. Leukemia 24(4):679–686
Chowdhury D et al (2008) A PP4-phosphatase complex dephosphorylates gamma-H2AX generated during DNA replication. Mol Cell 31(1):33–46
Lee DH et al (2010) A PP4 phosphatase complex dephosphorylates RPA2 to facilitate DNA repair via homologous recombination. Nat Struct Mol Biol 17(3):365–372
Pei H et al (2011) MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature 470(7332):124–128
Wang X et al (2001) Identification and characterization of a novel androgen receptor coregulator ARA267-alpha in prostate cancer cells. J Biol Chem 276(44):40417–40423
Toyokawa G et al (2011) Histone lysine methyltransferase Wolf–Hirschhorn syndrome candidate 1 is involved in human carcinogenesis through regulation of the Wnt pathway. Neoplasia 13(10):887–898
Gonzalez-Pecchi V et al (2020) NSD3S stabilizes MYC through hindering its interaction with FBXW7. J Mol Cell Biol 12(6):438–447
Pan C et al (2019) NSD1 mutations by HPV status in head and neck cancer: differences in survival and response to DNA-damaging agents. Cancers Head Neck 4(1):1–13
Peri S et al (2017) NSD1- and NSD2-damaging mutations define a subset of laryngeal tumors with favorable prognosis. Nat Commun 8(1):1–10
Bui N et al (2018) Disruption of NSD1 in head and neck cancer promotes favorable chemotherapeutic responses linked to hypomethylation. Mol Cancer Ther 17(7):1585–1594
Saloura V et al (2015) WHSC1 Promotes oncogenesis through regulation of NIMA-related kinase-7 in squamous cell carcinoma of the head and neck. Mol Cancer Res 13(2):293–304
Gameiro SF et al (2021) Low expression of NSD1, NSD2, and NSD3 define a subset of human papillomavirus-positive oral squamous carcinomas with unfavorable prognosis. Infect Agents Cancer 16(1):13
Ettel M et al (2019) Expression and prognostic value of NSD1 and SETD2 in pancreatic ductal adenocarcinoma and its precursor lesions. Pathology 51(4):392–398
Zhang S et al (2019) CRISPR/Cas9-mediated knockout of NSD1 suppresses the hepatocellular carcinoma development via the NSD1/H3/Wnt10b signaling pathway. J Exp Clin Cancer Res 38(1):1–14
Bianco-Miotto T et al (2010) Global levels of specific histone modifications and an epigenetic gene signature predict prostate cancer progression and development. Cancer Epidemiol Biomark Prev 19(10):2611–2622
Wang Y et al (2021) Histone methyltransferase WHSC1 inhibits colorectal cancer cell apoptosis via targeting anti-apoptotic BCL2. Cell Death Discov 7(1):1–9
Zhao L-H et al (2021) Identification of histone methyltransferase NSD2 as an important oncogenic gene in colorectal cancer. Cell Death Dis 12(11):974
He C et al (2019) Histone methyltransferase NSD2 regulates apoptosis and chemosensitivity in osteosarcoma. Cell Death Dis 10(2):1–13
Yang P et al (2012) Histone methyltransferase NSD2/MMSET mediates constitutive NF-B signaling for cancer cell proliferation, survival, and tumor growth via a feed-forward loop. Mol Cell Biol 32(15):3121–3131
Ezponda T et al (2013) The histone methyltransferase MMSET/WHSC1 activates TWIST1 to promote an epithelial–mesenchymal transition and invasive properties of prostate cancer. Oncogene 32(23):2882–2890
García-Carpizo V et al (2016) NSD2 contributes to oncogenic RAS-driven transcription in lung cancer cells through long-range epigenetic activation. Sci Rep 6(1):32952
Sengupta D et al (2021) NSD2 dimethylation at H3K36 promotes lung adenocarcinoma pathogenesis. Mol Cell 81(21):4481-4492.e9
Hudlebusch HR et al (2011) The histone methyltransferase and putative oncoprotein MMSET is overexpressed in a large variety of human tumors. Clin Cancer Res 17(9):2919–2933
Wang JJ et al (2019) Histone methyltransferase NSD2 mediates the survival and invasion of triple-negative breast cancer cells via stimulating ADAM9-EGFR-AKT signaling. Acta Pharmacol Sin 40(8):1067–1075
Hudlebusch HR et al (2011) MMSET is highly expressed and associated with aggressiveness in neuroblastoma. Cancer Res 71(12):4226–4235
Han X et al (2020) NSD2 promotes renal cancer progression through stimulating Akt/Erk signaling. Cancer Manag Res 12:375–383
Tonon G et al (2005) High-resolution genomic profiles of human lung cancer. Proc Natl Acad Sci U S A 102(27):9625–9630
Kassambara A, Klein B, Moreaux J (2009) MMSET is overexpressed in cancers: link with tumor aggressiveness. Biochem Biophys Res Commun 379(4):840–845
Berdasco M et al (2009) Epigenetic inactivation of the Sotos overgrowth syndrome gene histone methyltransferase NSD1 in human neuroblastoma and glioma. Proc Natl Acad Sci U S A 106(51):21830–21835
Su X et al (2017) NSD1 inactivation and SETD2 mutation drive a convergence toward loss of function of H3K36 writers in clear cell renal cell carcinomas. Cancer Res 77(18):4835–4845
Su X et al (2017) NSD1 inactivation and SETD2 mutation drive a convergence toward loss of function of H3K36 writers in clear cell renal cell carcinomas. Can Res 77(18):4835–4845
Yang ZQ et al (2010) Transforming properties of 8p11-12 amplified genes in human breast cancer. Cancer Res 70(21):8487–8497
Irish JC et al (2016) Amplification of WHSC1L1 regulates expression and estrogen-independent activation of ERα in SUM-44 breast cancer cells and is associated with ERα over-expression in breast cancer. Mol Oncol 10(6):850–865
Saloura V et al (2016) WHSC1L1 drives cell cycle progression through transcriptional regulation of CDC6 and CDK2 in squamous cell carcinoma of the head and neck. Oncotarget 7(27):42527–42538
Yuan G et al (2021) Elevated NSD3 histone methylation activity drives squamous cell lung cancer. Nature 590(7846):504–508
Yi L et al (2019) Downregulation of NSD3 (WHSC1L1) inhibits cell proliferation and migration via ERK1/2 deactivation and decreasing CAPG expression in colorectal cancer cells. Onco Targets Ther 12:3933–3943
Jeong GY et al (2021) NSD3-induced methylation of H3K36 activates NOTCH signaling to drive breast tumor initiation and metastatic progression. Cancer Res 81(1):77–90
D’Afonseca V et al (2020) Computational analyses on genetic alterations in the NSD genes family and the implications for colorectal cancer development. Ecancermedicalscience 14:1001
Kang D et al (2013) The histone methyltransferase Wolf–Hirschhorn syndrome candidate 1-like 1 (WHSC1L1) is involved in human carcinogenesis. Genes Chromosomes Cancer 52(2):126–139
French CA et al (2014) NSD3-NUT fusion oncoprotein in NUT midline carcinoma: implications for a novel oncogenic mechanism. Cancer Discov 4(8):928–941
Bauer DE et al (2012) Clinicopathologic features and long-term outcomes of NUT midline carcinoma. Clin Cancer Res 18(20):5773–5779
Agaimy A et al (2021) Misleading germ cell phenotype in pulmonary NUT carcinoma harboring the ZNF532-NUTM1 fusion. Am J Surg Pathol 46:281–288
Stevens TM et al (2019) NUTM1-rearranged neoplasia: a multi-institution experience yields novel fusion partners and expands the histologic spectrum. Mod Pathol 32(6):764–773
Chau NG et al (2020) An anatomical site and genetic-based prognostic model for patients with nuclear protein in testis (NUT) midline carcinoma: analysis of 124 patients. JNCI Cancer Spectr 4(2):pkz094
Zhao X et al (2019) Downregulation of MMSET impairs breast cancer proliferation and metastasis through inhibiting Wnt/β-catenin signaling. Onco Targets Ther 12:1965–1977
Huang Z et al (2013) NSD2 is recruited through its PHD domain to oncogenic gene loci to drive multiple myeloma. Cancer Res 73(20):6277–6288
Kang HB et al (2009) The histone methyltransferase, NSD2, enhances androgen receptor-mediated transcription. FEBS Lett 583(12):1880–1886
Sun Y et al (2021) Elevated expression of nuclear receptor-binding SET domain 3 promotes pancreatic cancer cell growth. Cell Death Dis 12(10):913
Mahmood SF et al (2013) PPAPDC1B and WHSC1L1 are common drivers of the 8p11-12 amplicon, not only in breast tumors but also in pancreatic adenocarcinomas and lung tumors. Am J Pathol 183(5):1634–1644
Rahman S et al (2011) The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol Cell Biol 31(13):2641–2652
Saloura V et al (2017) WHSC1L1-mediated EGFR mono-methylation enhances the cytoplasmic and nuclear oncogenic activity of EGFR in head and neck cancer. Sci Rep 7:40664
Song D et al (2021) NSD2 promotes tumor angiogenesis through methylating and activating STAT3 protein. Oncogene 40(16):2952–2967
Liu Z et al (2017) Silencing of histone methyltransferase NSD3 reduces cell viability in osteosarcoma with induction of apoptosis. Oncol Rep 38(5):2796–2802
Aytes A et al (2018) NSD2 is a conserved driver of metastatic prostate cancer progression. Nat Commun 9(1):5201
Yuan S et al (2020) Global regulation of the histone mark H3K36me2 underlies epithelial plasticity and metastatic progression. Cancer Discov 10(6):854–871
Han X et al (2019) Knockdown of NSD2 suppresses renal cell carcinoma metastasis by inhibiting epithelial–mesenchymal transition. Int J Med Sci 16(10):1404–1411
Lu T et al (2010) Regulation of NF-B by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc Natl Acad Sci 107(1):46–51
Kudithipudi S et al (2014) Substrate specificity analysis and novel substrates of the protein lysine methyltransferase NSD1. Chem Biol 21(2):226–237
Murciano-Goroff YR, Warner AB, Wolchok JD (2020) The future of cancer immunotherapy: microenvironment-targeting combinations. Cell Res 30(6):507–519
Brennan K et al (2017) NSD1 inactivation defines an immune cold, DNA hypomethylated subtype in squamous cell carcinoma. Sci Rep 7(1):1–12
Saloura V et al (2019) Immune profiles in primary squamous cell carcinoma of the head and neck. Oral Oncol 96:77–88
Want MY et al (2021) WHSC1/NSD2 regulates immune infiltration in prostate cancer. J Immunother Cancer 9(2):e001374
Cedar H, Bergman Y (2009) Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet 10(5):295–304
Baubec T et al (2015) Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature 520(7546):243–247
Pangeni RP et al (2020) G9a regulates tumorigenicity and stemness through genome-wide DNA methylation reprogramming in non-small cell lung cancer. Clin Epigenet 12(1):88
Zhang K et al (2018) Targeting histone methyltransferase G9a inhibits growth and Wnt signaling pathway by epigenetically regulating HP1alpha and APC2 gene expression in non-small cell lung cancer. Mol Cancer 17(1):153
Li H et al (2006) The histone methyltransferase SETDB1 and the DNA methyltransferase DNMT3A interact directly and localize to promoters silenced in cancer cells. J Biol Chem 281(28):19489–19500
Castillo-Aguilera O et al (2017) DNA methylation targeting: the DNMT/HMT crosstalk challenge. Biomolecules 7(1):3
Neri F et al (2017) Intragenic DNA methylation prevents spurious transcription initiation. Nature 543(7643):72–77
Lee ST, Wiemels JL (2016) Genome-wide CpG island methylation and intergenic demethylation propensities vary among different tumor sites. Nucleic Acids Res 44(3):1105–1117
Choufani S et al (2015) NSD1 mutations generate a genome-wide DNA methylation signature. Nat Commun 6:10207
Cancer Genome Atlas Network (2015) Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 517(7536):576–582
Brennan K et al (2017) Identification of an atypical etiological head and neck squamous carcinoma subtype featuring the CpG island methylator phenotype. EBioMedicine 17:223–236
Suzuki S, Murakami Y, Takahata S (2017) H3K36 methylation state and associated silencing mechanisms. Transcription 8(1):26–31
DiFiore JV et al (2020) Unique and shared roles for histone H3K36 methylation states in transcription regulation functions. Cell Rep 31(10):107751
O’Hagan HM, Mohammad HP, Baylin SB (2008) Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genet 4(8):e1000155
Mortusewicz O et al (2005) Recruitment of DNA methyltransferase I to DNA repair sites. Proc Natl Acad Sci U S A 102(25):8905–8909
Xue W et al (2021) Long non-coding RNAs MACC1-AS1 and FOXD2-AS1 mediate NSD2-induced cisplatin resistance in esophageal squamous cell carcinoma. Mol Ther Nucleic Acids 23:592–602
Wei J et al (2014) Differential effect of MMSET mRNA levels on survival to first-line FOLFOX and second-line docetaxel in gastric cancer. Br J Cancer 110(11):2662–2668
Dawson MA, Kouzarides T (2012) Cancer epigenetics: from mechanism to therapy. Cell 150(1):12–27
Huang H et al (2020) Covalent inhibition of NSD1 histone methyltransferase. Nat Chem Biol 16(12):1403–1410
Kubicek S et al (2007) Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell 25(3):473–481
Morishita M et al (2017) BIX-01294 inhibits oncoproteins NSD1, NSD2 and NSD3. Med Chem Res 26(9):2038–2047
di Luccio E (2015) Inhibition of nuclear receptor binding SET domain 2/multiple myeloma SET domain by LEM-06 implication for epigenetic cancer therapies. J Cancer Prev 20(2):113–120
Shen Y et al (2019) Identification of LEM-14 inhibitor of the oncoprotein NSD2. Biochem Biophys Res Commun 508(1):102–108
Coussens NP et al (2018) High-throughput screening with nucleosome substrate identifies small-molecule inhibitors of the human histone lysine methyltransferase NSD2. J Biol Chem 293(35):13750–13765
Morrison MJ et al (2018) Identification of a peptide inhibitor for the histone methyltransferase WHSC1. PLoS One 13(5):e0197082
Qin S, Min J (2014) Structure and function of the nucleosome-binding PWWP domain. Trends Biochem Sci 39(11):536–547
Vermeulen M et al (2010) Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 142(6):967–980
Böttcher J et al (2019) Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3. Nat Chem Biol 15(8):822–829
Dilworth D et al (2022) A chemical probe targeting the PWWP domain alters NSD2 nucleolar localization. Nat Chem Biol 18(1):56–63
Funding
NIH R01 CA218802 grant, and the NCI NIH Core Grant P30 CA060553 to the Robert H Lurie Comprehensive Cancer Center at Northwestern University, Translational Bridge Award from Northwestern University (to Y.B.). This study was also partially supported by 1P50 DE030707-01 and USAMRAA W81XWH2110487 (to E.G.), NIH/NCI R01CA227918 (to J.Y.) Department of Defense grants W81XWH-17-1-0405 and W81XWH-17-1-0578 (to J.Y.), R01DE027809 to E. I., and Prostate Cancer Foundation 2017CHAL2008 (to J.Y.); J.K., and E.G. were in part supported by the NCI Core Grant P30 CA006927 (to Fox Chase Cancer Center).
Author information
Authors and Affiliations
Contributions
IT, RP and IB conducted the literature review and wrote the first draft. IT, RP and SM prepared the figures and tables. JK, JY, EI, EG, and YB conducted reviewing proofreading and content processing.
Corresponding author
Ethics declarations
Conflict of interest
The author declares he has no conflict of interest.
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Topchu, I., Pangeni, R.P., Bychkov, I. et al. The role of NSD1, NSD2, and NSD3 histone methyltransferases in solid tumors. Cell. Mol. Life Sci. 79, 285 (2022). https://doi.org/10.1007/s00018-022-04321-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-022-04321-2