Abstract
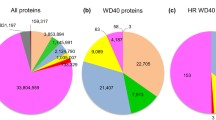
The superfamily of armadillo repeat proteins is a fascinating archetype of modular-binding proteins involved in various fundamental cellular processes, including cell–cell adhesion, cytoskeletal organization, nuclear import, and molecular signaling. Despite their diverse functions, they all share tandem armadillo (ARM) repeats, which stack together to form a conserved three-dimensional structure. This superhelical armadillo structure enables them to interact with distinct partners by wrapping around them. Despite the important functional roles of this superfamily, a comprehensive analysis of the composition, classification, and phylogeny of this protein superfamily has not been reported. Furthermore, relatively little is known about a subset of ARM proteins, and some of the current annotations of armadillo repeats are incomplete or incorrect, often due to high similarity with HEAT repeats. We identified the entire armadillo repeat superfamily repertoire in the human genome, annotated each armadillo repeat, and performed an extensive evolutionary analysis of the armadillo repeat proteins in both metazoan and premetazoan species. Phylogenetic analyses of the superfamily classified them into several discrete branches with members showing significant sequence homology, and often also related functions. Interestingly, the phylogenetic structure of the superfamily revealed that about 30 % of the members predate metazoans and represent an ancient subset, which is gradually evolving to acquire complex and highly diverse functions.



Similar content being viewed by others
References
Riggleman B, Wieschaus E, Schedl P (1989) Molecular analysis of the armadillo locus: uniformly distributed transcripts and a protein with novel internal repeats are associated with a Drosophila segment polarity gene. Genes Dev 3(1):96–113
Peifer M, Berg S, Reynolds AB (1994) A repeating amino acid motif shared by proteins with diverse cellular roles. Cell 76(5):789–791
Tewari R, Bailes E, Bunting KA, Coates JC (2010) Armadillo-repeat protein functions: questions for little creatures. Trends Cell Biol 20(8):470–481. doi:10.1016/j.tcb.2010.05.003
Huber AH, Nelson WJ, Weis WI (1997) Three-dimensional structure of the armadillo repeat region of beta-catenin. Cell 90(5):871–882
Madhurantakam C, Varadamsetty G, Grutter MG, Pluckthun A, Mittl PR (2012) Structure-based optimization of designed Armadillo-repeat proteins. Protein Sci 21(7):1015–1028. doi:10.1002/pro.2085
Parmeggiani F, Pellarin R, Larsen AP, Varadamsetty G, Stumpp MT, Zerbe O, Caflisch A, Pluckthun A (2008) Designed armadillo repeat proteins as general peptide-binding scaffolds: consensus design and computational optimization of the hydrophobic core. J Mol Biol 376(5):1282–1304. doi:10.1016/j.jmb.2007.12.014
Cingolani G, Petosa C, Weis K, Muller CW (1999) Structure of importin-beta bound to the IBB domain of importin-alpha. Nature 399(6733):221–229. doi:10.1038/20367
Choi HJ, Gross JC, Pokutta S, Weis WI (2009) Interactions of plakoglobin and beta-catenin with desmosomal cadherins: basis of selective exclusion of alpha- and beta-catenin from desmosomes. J Biol Chem 284(46):31776–31788. doi:10.1074/jbc.M109.047928
Ishiyama N, Lee SH, Liu S, Li GY, Smith MJ, Reichardt LF, Ikura M (2010) Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell–cell adhesion. Cell 141(1):117–128. doi:10.1016/j.cell.2010.01.017
Andrade MA, Perez-Iratxeta C, Ponting CP (2001) Protein repeats: structures, functions, and evolution. J Struct Biol 134(2–3):117–131. doi:10.1006/jsbi.2001.4392
Kippert F, Gerloff DL (2009) Highly sensitive detection of individual HEAT and ARM repeats with HHpred and COACH. PLoS One 4(9):e7148. doi:10.1371/journal.pone.0007148
Kuhn S, Erdmann C, Kage F, Block J, Schwenkmezger L, Steffen A, Rottner K, Geyer M (2015) The structure of FMNL2-Cdc42 yields insights into the mechanism of lamellipodia and filopodia formation. Nat Commun 6:7088. doi:10.1038/ncomms8088
Rose R, Weyand M, Lammers M, Ishizaki T, Ahmadian MR, Wittinghofer A (2005) Structural and mechanistic insights into the interaction between Rho and mammalian Dia. Nature 435(7041):513–518. doi:10.1038/nature03604
Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, Holm L, Sonnhammer EL, Eddy SR, Bateman A (2010) The Pfam protein families database. Nucleic Acids Res 38(Database issue):D211–D222. doi:10.1093/nar/gkp985
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32(5):1792–1797. doi:10.1093/nar/gkh340
Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH (2015) CDD: NCBI’s conserved domain database. Nucleic Acids Res 43(Database issue):D222–D226. doi:10.1093/nar/gku1221
Dalquen DA, Dessimoz C (2013) Bidirectional best hits miss many orthologs in duplication-rich clades such as plants and animals. Genome Biol Evol 5(10):1800–1806. doi:10.1093/gbe/evt132
Solovyev V, Kosarev P, Seledsov I, Vorobyev D (2006) Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol 7(Suppl 1):S10 11–12. doi:10.1186/gb-2006-7-s1-s10
Burge CB, Karlin S (1998) Finding the genes in genomic DNA. Curr Opin Struct Biol 8(3):346–354
Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL (2008) NCBI BLAST: a better web interface. Nucleic Acids Res 36(Web Server issue):W5–W9. doi:10.1093/nar/gkn201
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19(12):1572–1574
Letunic I, Bork P (2007) Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23(1):127–128. doi:10.1093/bioinformatics/btl529
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. doi:10.1093/bioinformatics/btu033
Pruitt KD, Brown GR, Hiatt SM, Thibaud-Nissen F, Astashyn A, Ermolaeva O, Farrell CM, Hart J, Landrum MJ, McGarvey KM, Murphy MR, O’Leary NA, Pujar S, Rajput B, Rangwala SH, Riddick LD, Shkeda A, Sun H, Tamez P, Tully RE, Wallin C, Webb D, Weber J, Wu W, DiCuccio M, Kitts P, Maglott DR, Murphy TD, Ostell JM (2014) RefSeq: an update on mammalian reference sequences. Nucleic Acids Res 42(Database issue):D756–D763. doi:10.1093/nar/gkt1114
Imasaki T, Shimizu T, Hashimoto H, Hidaka Y, Kose S, Imamoto N, Yamada M, Sato M (2007) Structural basis for substrate recognition and dissociation by human transportin 1. Mol Cell 28(1):57–67. doi:10.1016/j.molcel.2007.08.006
Mitchell A, Chang HY, Daugherty L, Fraser M, Hunter S, Lopez R, McAnulla C, McMenamin C, Nuka G, Pesseat S, Sangrador-Vegas A, Scheremetjew M, Rato C, Yong SY, Bateman A, Punta M, Attwood TK, Sigrist CJ, Redaschi N, Rivoire C, Xenarios I, Kahn D, Guyot D, Bork P, Letunic I, Gough J, Oates M, Haft D, Huang H, Natale DA, Wu CH, Orengo C, Sillitoe I, Mi H, Thomas PD, Finn RD (2015) The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res 43(Database issue):D213–D221. doi:10.1093/nar/gku1243
Suga H, Chen Z, de Mendoza A, Sebe-Pedros A, Brown MW, Kramer E, Carr M, Kerner P, Vervoort M, Sanchez-Pons N, Torruella G, Derelle R, Manning G, Lang BF, Russ C, Haas BJ, Roger AJ, Nusbaum C, Ruiz-Trillo I (2013) The Capsaspora genome reveals a complex unicellular prehistory of animals. Nat Commun 4:2325. doi:10.1038/ncomms3325
Goldfarb DS, Corbett AH, Mason DA, Harreman MT, Adam SA (2004) Importin alpha: a multipurpose nuclear-transport receptor. Trends Cell Biol 14(9):505–514. doi:10.1016/j.tcb.2004.07.016
Chalkia D, Nikolaidis N, Makalowski W, Klein J, Nei M (2008) Origins and evolution of the formin multigene family that is involved in the formation of actin filaments. Mol Biol Evol 25(12):2717–2733. doi:10.1093/molbev/msn215
Lopez-Domenech G, Serrat R, Mirra S, D’Aniello S, Somorjai I, Abad A, Vitureira N, Garcia-Arumi E, Alonso MT, Rodriguez-Prados M, Burgaya F, Andreu AL, Garcia-Sancho J, Trullas R, Garcia-Fernandez J, Soriano E (2012) The Eutherian Armcx genes regulate mitochondrial trafficking in neurons and interact with Miro and Trak2. Nat Commun 3:814. doi:10.1038/ncomms1829
Zhao ZM, Reynolds AB, Gaucher EA (2011) The evolutionary history of the catenin gene family during metazoan evolution. BMC Evol Biol 11:198. doi:10.1186/1471-2148-11-198
Hall BG (2005) Comparison of the accuracies of several phylogenetic methods using protein and DNA sequences. Mol Biol Evol 22(3):792–802. doi:10.1093/molbev/msi066
Gaucher EA, Kratzer JT, Randall RN (2010) Deep phylogeny—how a tree can help characterize early life on Earth. Cold Spring Harb Perspect Biol 2(1):a002238. doi:10.1101/cshperspect.a002238
Wang LS, Leebens-Mack J, Kerr Wall P, Beckmann K, dePamphilis CW, Warnow T (2011) The impact of multiple protein sequence alignment on phylogenetic estimation. IEEE/ACM Trans Comput Biol Bioinform 8(4):1108–1119. doi:10.1109/TCBB.2009.68
Garces RG, Gillon W, Pai EF (2007) Atomic model of human Rcd-1 reveals an armadillo-like-repeat protein with in vitro nucleic acid binding properties. Protein Sci 16(2):176–188. doi:10.1110/ps.062600507
Schonichen A (1803) Geyer M (2010) Fifteen formins for an actin filament: a molecular view on the regulation of human formins. Biochim Biophys Acta 2:152–163. doi:10.1016/j.bbamcr.2010.01.014
Hulpiau P, Gul IS, van Roy F (2013) New insights into the evolution of metazoan cadherins and catenins. Prog Mol Biol Transl Sci 116:71–94. doi:10.1016/B978-0-12-394311-8.00004-2
Hulpiau P, van Roy F (2011) New insights into the evolution of metazoan cadherins. Mol Biol Evol 28(1):647–657. doi:10.1093/molbev/msq233
Yamamoto Y, Izumi Y, Matsuzaki F (2008) The GC kinase Fray and Mo25 regulate Drosophila asymmetric divisions. Biochem Biophys Res Commun 366(1):212–218. doi:10.1016/j.bbrc.2007.11.128
Filippi BM, de los Heros P, Mehellou Y, Navratilova I, Gourlay R, Deak M, Plater L, Toth R, Zeqiraj E, Alessi DR (2011) MO25 is a master regulator of SPAK/OSR1 and MST3/MST4/YSK1 protein kinases. EMBO J 30(9):1730–1741. doi:10.1038/emboj.2011.78
Mendoza M, Redemann S, Brunner D (2005) The fission yeast MO25 protein functions in polar growth and cell separation. Eur J Cell Biol 84(12):915–926. doi:10.1016/j.ejcb.2005.09.013
Suzuki T, Ueda A, Kobayashi N, Yang J, Tomaru K, Yamamoto M, Takeno M, Ishigatsubo Y (2008) Proteasome-dependent degradation of alpha-catenin is regulated by interaction with ARMc8alpha. Biochem J 411(3):581–591. doi:10.1042/BJ20071312
Menssen R, Schweiggert J, Schreiner J, Kusevic D, Reuther J, Braun B, Wolf DH (2012) Exploring the topology of the Gid complex, the E3 ubiquitin ligase involved in catabolite-induced degradation of gluconeogenic enzymes. J Biol Chem 287(30):25602–25614. doi:10.1074/jbc.M112.363762
Francis O, Han F, Adams JC (2013) Molecular phylogeny of a RING E3 ubiquitin ligase, conserved in eukaryotic cells and dominated by homologous components, the muskelin/RanBPM/CTLH complex. PLoS One 8(10):e75217. doi:10.1371/journal.pone.0075217
Nakajima H, Hirata A, Ogawa Y, Yonehara T, Yoda K, Yamasaki M (1991) A cytoskeleton-related gene, uso1, is required for intracellular protein transport in Saccharomyces cerevisiae. J Cell Biol 113(2):245–260
Striegl H, Roske Y, Kummel D, Heinemann U (2009) Unusual armadillo fold in the human general vesicular transport factor p115. PLoS One 4(2):e4656. doi:10.1371/journal.pone.0004656
Geles KG, Adam SA (2001) Germline and developmental roles of the nuclear transport factor importin alpha3 in C. elegans. Development 128(10):1817–1830
Phadnis N, Hsieh E, Malik HS (2012) Birth, death, and replacement of karyopherins in Drosophila. Mol Biol Evol 29(5):1429–1440. doi:10.1093/molbev/msr306
Grimson MJ, Coates JC, Reynolds JP, Shipman M, Blanton RL, Harwood AJ (2000) Adherens junctions and beta-catenin-mediated cell signalling in a non-metazoan organism. Nature 408(6813):727–731. doi:10.1038/35047099
Coates JC, Grimson MJ, Williams RS, Bergman W, Blanton RL, Harwood AJ (2002) Loss of the beta-catenin homologue aardvark causes ectopic stalk formation in Dictyostelium. Mech Dev 116(1–2):117–127
Dickinson DJ, Nelson WJ, Weis WI (2011) A polarized epithelium organized by beta- and alpha-catenin predates cadherin and metazoan origins. Science 331(6022):1336–1339. doi:10.1126/science.1199633
Ha NC, Tonozuka T, Stamos JL, Choi HJ, Weis WI (2004) Mechanism of phosphorylation-dependent binding of APC to beta-catenin and its role in beta-catenin degradation. Mol Cell 15(4):511–521. doi:10.1016/j.molcel.2004.08.010
Yang J, Zhang W, Evans PM, Chen X, He X, Liu C (2006) Adenomatous polyposis coli (APC) differentially regulates beta-catenin phosphorylation and ubiquitination in colon cancer cells. J Biol Chem 281(26):17751–17757. doi:10.1074/jbc.M600831200
Cai X, Zhang Y (2006) Molecular evolution of the ankyrin gene family. Mol Biol Evol 23(3):550–558. doi:10.1093/molbev/msj056
Abu-Helo A, Simonin F (2010) Identification and biological significance of G protein-coupled receptor associated sorting proteins (GASPs). Pharmacol Ther 126(3):244–250. doi:10.1016/j.pharmthera.2010.03.004
Simonin F, Karcher P, Boeuf JJ, Matifas A, Kieffer BL (2004) Identification of a novel family of G protein-coupled receptor associated sorting proteins. J Neurochem 89(3):766–775. doi:10.1111/j.1471-4159.2004.02411.x
Kemphues KJ, Wolf N, Wood WB, Hirsh D (1986) Two loci required for cytoplasmic organization in early embryos of Caenorhabditis elegans. Dev Biol 113(2):449–460
Reichen C, Hansen S, Pluckthun A (2014) Modular peptide binding: from a comparison of natural binders to designed armadillo repeat proteins. J Struct Biol 185(2):147–162. doi:10.1016/j.jsb.2013.07.012
Stamos JL, Weis WI (2013) The beta-catenin destruction complex. Cold Spring Harb Perspect Biol 5(1). doi:10.1101/cshperspect.a007898
Hatzfeld M (2007) Plakophilins: multifunctional proteins or just regulators of desmosomal adhesion? Biochim Biophys Acta 1773(1):69–77. doi:10.1016/j.bbamcr.2006.04.009
Carramusa L, Ballestrem C, Zilberman Y, Bershadsky AD (2007) Mammalian diaphanous-related formin Dia1 controls the organization of E-cadherin-mediated cell–cell junctions. J Cell Sci 120(Pt 21):3870–3882. doi:10.1242/jcs.014365
Ireton RC, Davis MA, van Hengel J, Mariner DJ, Barnes K, Thoreson MA, Anastasiadis PZ, Matrisian L, Bundy LM, Sealy L, Gilbert B, van Roy F, Reynolds AB (2002) A novel role for p120 catenin in E-cadherin function. J Cell Biol 159(3):465–476. doi:10.1083/jcb.200205115
Kourtidis A, Ngok SP, Anastasiadis PZ (2013) p120 catenin: an essential regulator of cadherin stability, adhesion-induced signaling, and cancer progression. Prog Mol Biol Transl Sci 116:409–432. doi:10.1016/B978-0-12-394311-8.00018-2
Young KG (1803) Copeland JW (2010) Formins in cell signaling. Biochim Biophys Acta 2:183–190. doi:10.1016/j.bbamcr.2008.09.017
Buckley CD, Tan J, Anderson KL, Hanein D, Volkmann N, Weis WI, Nelson WJ, Dunn AR (2014) Cell adhesion. The minimal cadherin-catenin complex binds to actin filaments under force. Science 346(6209):1254211. doi:10.1126/science.1254211
Valenta T, Hausmann G, Basler K (2012) The many faces and functions of beta-catenin. EMBO J 31(12):2714–2736. doi:10.1038/emboj.2012.150
Haraguchi K, Hayashi T, Jimbo T, Yamamoto T, Akiyama T (2006) Role of the kinesin-2 family protein, KIF3, during mitosis. J Biol Chem 281(7):4094–4099. doi:10.1074/jbc.M507028200
Kaplan KB, Burds AA, Swedlow JR, Bekir SS, Sorger PK, Nathke IS (2001) A role for the adenomatous polyposis coli protein in chromosome segregation. Nat Cell Biol 3(4):429–432. doi:10.1038/35070123
Stolz A, Neufeld K, Ertych N, Bastians H (2015) Wnt-mediated protein stabilization ensures proper mitotic microtubule assembly and chromosome segregation. EMBO Rep 16(4):490–499. doi:10.15252/embr.201439410
Jimbo T, Kawasaki Y, Koyama R, Sato R, Takada S, Haraguchi K, Akiyama T (2002) Identification of a link between the tumour suppressor APC and the kinesin superfamily. Nat Cell Biol 4(4):323–327. doi:10.1038/ncb779
Onoufriadis A, Shoemark A, Munye MM, James CT, Schmidts M, Patel M, Rosser EM, Bacchelli C, Beales PL, Scambler PJ, Hart SL, Danke-Roelse JE, Sloper JJ, Hull S, Hogg C, Emes RD, Pals G, Moore AT, Chung EM, UK10 K, Mitchison HM (2014) Combined exome and whole-genome sequencing identifies mutations in ARMC4 as a cause of primary ciliary dyskinesia with defects in the outer dynein arm. J Med Genet 51(1):61–67. doi:10.1136/jmedgenet-2013-101938
Cheng W, Ip YT, Xu Z (2013) Gudu, an Armadillo repeat-containing protein, is required for spermatogenesis in Drosophila. Gene 531(2):294–300. doi:10.1016/j.gene.2013.08.080
Pausch H, Venhoranta H, Wurmser C, Hakala K, Iso-Touru T, Sironen A, Vingborg RK, Lohi H, Soderquist L, Fries R, Andersson M (2016) A frameshift mutation in ARMC3 is associated with a tail stump sperm defect in Swedish Red (Bos taurus) cattle. BMC Genet 17(1):49. doi:10.1186/s12863-016-0356-7
Iida H, Urasoko A, Doiguchi M, Mori T, Toshimori K, Shibata Y (2003) Complementary DNA cloning and characterization of rat spergen-2, a spermatogenic cell-specific gene 2 encoding a 56-kilodalton nuclear protein bearing ankyrin repeat motifs. Biol Reprod 69(2):421–429. doi:10.1095/biolreprod.102.013987
Wang HL, Fan SS, Pang M, Liu YH, Guo M, Liang JB, Zhang JL, Yu BF, Guo R, Xie J, Zheng GP (2015) The Ankyrin repeat domain 49 (ANKRD49) augments autophagy of serum-starved GC-1 cells through the NF-kappaB pathway. PLoS One 10(6):e0128551. doi:10.1371/journal.pone.0128551
Straschil U, Talman AM, Ferguson DJ, Bunting KA, Xu Z, Bailes E, Sinden RE, Holder AA, Smith EF, Coates JC, Rita T (2010) The Armadillo repeat protein PF16 is essential for flagellar structure and function in Plasmodium male gametes. PLoS One 5(9):e12901. doi:10.1371/journal.pone.0012901
Holt JE, Ly-Huynh JD, Efthymiadis A, Hime GR, Loveland KL, Jans DA (2007) Regulation of nuclear import during differentiation; the IMP alpha gene family and spermatogenesis. Curr Genomics 8(5):323–334. doi:10.2174/138920207782446151
Mudgil Y, Shiu SH, Stone SL, Salt JN, Goring DR (2004) A large complement of the predicted Arabidopsis ARM repeat proteins are members of the U-box E3 ubiquitin ligase family. Plant Physiol 134(1):59–66. doi:10.1104/pp.103.029553
Sharma M, Singh A, Shankar A, Pandey A, Baranwal V, Kapoor S, Tyagi AK, Pandey GK (2014) Comprehensive expression analysis of rice Armadillo gene family during abiotic stress and development. DNA Res 21(3):267–283. doi:10.1093/dnares/dst056
Teotia S, Lamb RS (2011) RCD1 and SRO1 are necessary to maintain meristematic fate in Arabidopsis thaliana. J Exp Bot 62(3):1271–1284. doi:10.1093/jxb/erq363
Latijnhouwers M, Gillespie T, Boevink P, Kriechbaumer V, Hawes C, Carvalho CM (2007) Localization and domain characterization of Arabidopsis golgin candidates. J Exp Bot 58(15–16):4373–4386. doi:10.1093/jxb/erm304
Baumgartner W (2013) Possible roles of LI-Cadherin in the formation and maintenance of the intestinal epithelial barrier. Tissue Barriers 1(1):e23815. doi:10.4161/tisb.23815
Oda H, Takeichi M (2011) Evolution: structural and functional diversity of cadherin at the adherens junction. J Cell Biol 193(7):1137–1146. doi:10.1083/jcb.201008173
Sotomayor M, Gaudet R, Corey DP (2014) Sorting out a promiscuous superfamily: towards cadherin connectomics. Trends Cell Biol 24(9):524–536. doi:10.1016/j.tcb.2014.03.007
Xing Y, Takemaru K, Liu J, Berndt JD, Zheng JJ, Moon RT, Xu W (2008) Crystal structure of a full-length beta-catenin. Structure 16(3):478–487. doi:10.1016/j.str.2007.12.021
Schuld NJ, Hauser AD, Gastonguay AJ, Wilson JM, Lorimer EL, Williams CL (2014) SmgGDS-558 regulates the cell cycle in pancreatic, non-small cell lung, and breast cancers. Cell Cycle 13(6):941–952. doi:10.4161/cc.27804
Hauser AD, Bergom C, Schuld NJ, Chen X, Lorimer EL, Huang J, Mackinnon AC, Williams CL (2014) The SmgGDS splice variant SmgGDS-558 is a key promoter of tumor growth and RhoA signaling in breast cancer. Mol Cancer Res 12(1):130–142. doi:10.1158/1541-7786.MCR-13-0362
Tew GW, Lorimer EL, Berg TJ, Zhi H, Li R, Williams CL (2008) SmgGDS regulates cell proliferation, migration, and NF-kappaB transcriptional activity in non-small cell lung carcinoma. J Biol Chem 283(2):963–976. doi:10.1074/jbc.M707526200
de Bruyn KM, Zwartkruis FJ, de Rooij J, Akkerman JW, Bos JL (2003) The small GTPase Rap1 is activated by turbulence and is involved in integrin [alpha]IIb[beta]3-mediated cell adhesion in human megakaryocytes. J Biol Chem 278(25):22412–22417. doi:10.1074/jbc.M212036200
Chung KT, Shen Y, Hendershot LM (2002) BAP, a mammalian BiP-associated protein, is a nucleotide exchange factor that regulates the ATPase activity of BiP. J Biol Chem 277(49):47557–47563. doi:10.1074/jbc.M208377200
Inaguma Y, Hamada N, Tabata H, Iwamoto I, Mizuno M, Nishimura YV, Ito H, Morishita R, Suzuki M, Ohno K, Kumagai T, Nagata K (2014) SIL1, a causative cochaperone gene of Marinesco–Sojgren syndrome, plays an essential role in establishing the architecture of the developing cerebral cortex. EMBO Mol Med 6(3):414–429. doi:10.1002/emmm.201303069
Lee CF, Hauenstein AV, Fleming JK, Gasper WC, Engelke V, Sankaran B, Bernstein SI, Huxford T (2011) X-ray crystal structure of the UCS domain-containing UNC-45 myosin chaperone from Drosophila melanogaster. Structure 19(3):397–408. doi:10.1016/j.str.2011.01.002
Price MG, Landsverk ML, Barral JM, Epstein HF (2002) Two mammalian UNC-45 isoforms are related to distinct cytoskeletal and muscle-specific functions. J Cell Sci 115(Pt 21):4013–4023
Jilani Y, Lu S, Lei H, Karnitz LM, Chadli A (2015) UNC45A localizes to centrosomes and regulates cancer cell proliferation through ChK1 activation. Cancer Lett 357(1):114–120. doi:10.1016/j.canlet.2014.11.009
Nozaki M, Onishi Y, Togashi S, Miyamoto H (1996) Molecular characterization of the Drosophila Mo25 gene, which is conserved among Drosophila, mouse, and yeast. DNA Cell Biol 15(6):505–509
Zeqiraj E, Filippi BM, Deak M, Alessi DR, van Aalten DM (2009) Structure of the LKB1-STRAD-MO25 complex reveals an allosteric mechanism of kinase activation. Science 326(5960):1707–1711. doi:10.1126/science.1178377
Dettmann A, Illgen J, Marz S, Schurg T, Fleissner A, Seiler S (2012) The NDR kinase scaffold HYM1/MO25 is essential for MAK2 map kinase signaling in Neurospora crassa. PLoS Genet 8(9):e1002950. doi:10.1371/journal.pgen.1002950
Alberti S, Bohse K, Arndt V, Schmitz A, Hohfeld J (2004) The cochaperone HspBP1 inhibits the CHIP ubiquitin ligase and stimulates the maturation of the cystic fibrosis transmembrane conductance regulator. Mol Biol Cell 15(9):4003–4010. doi:10.1091/mbc.E04-04-0293
Rogon C, Ulbricht A, Hesse M, Alberti S, Vijayaraj P, Best D, Adams IR, Magin TM, Fleischmann BK, Hohfeld J (2014) HSP70-binding protein HSPBP1 regulates chaperone expression at a posttranslational level and is essential for spermatogenesis. Mol Biol Cell 25(15):2260–2271. doi:10.1091/mbc.E14-02-0742
Feral C, Wu YQ, Pawlak A, Guellaen G (2001) Meiotic human sperm cells express a leucine-rich homologue of Caenorhabditis elegans early embryogenesis gene, Zyg-11. Mol Hum Reprod 7(12):1115–1122
Vasudevan S, Starostina NG, Kipreos ET (2007) The Caenorhabditis elegans cell-cycle regulator ZYG-11 defines a conserved family of CUL-2 complex components. EMBO Rep 8(3):279–286. doi:10.1038/sj.embor.7400895
Hjeij R, Lindstrand A, Francis R, Zariwala MA, Liu X, Li Y, Damerla R, Dougherty GW, Abouhamed M, Olbrich H, Loges NT, Pennekamp P, Davis EE, Carvalho CM, Pehlivan D, Werner C, Raidt J, Kohler G, Haffner K, Reyes-Mugica M, Lupski JR, Leigh MW, Rosenfeld M, Morgan LC, Knowles MR, Lo CW, Katsanis N, Omran H (2013) ARMC4 mutations cause primary ciliary dyskinesia with randomization of left/right body asymmetry. Am J Hum Genet 93(2):357–367. doi:10.1016/j.ajhg.2013.06.009
Lonergan KM, Chari R, Deleeuw RJ, Shadeo A, Chi B, Tsao MS, Jones S, Marra M, Ling V, Ng R, Macaulay C, Lam S, Lam WL (2006) Identification of novel lung genes in bronchial epithelium by serial analysis of gene expression. Am J Respir Cell Mol Biol 35(6):651–661. doi:10.1165/rcmb.2006-0056OC
Sapiro R, Kostetskii I, Olds-Clarke P, Gerton GL, Radice GL, Strauss IJ (2002) Male infertility, impaired sperm motility, and hydrocephalus in mice deficient in sperm-associated antigen 6. Mol Cell Biol 22(17):6298–6305
Teves ME, Sears PR, Li W, Zhang Z, Tang W, van Reesema L, Costanzo RM, Davis CW, Knowles MR, Strauss JF 3rd, Zhang Z (2014) Sperm-associated antigen 6 (SPAG6) deficiency and defects in ciliogenesis and cilia function: polarity, density, and beat. PLoS One 9(10):e107271. doi:10.1371/journal.pone.0107271
Li W, Mukherjee A, Wu J, Zhang L, Teves ME, Li H, Nambiar S, Henderson SC, Horwitz AR, Strauss Iii JF, Fang X, Zhang Z (2015) Sperm Associated Antigen 6 (SPAG6) regulates fibroblast cell growth, morphology, migration and ciliogenesis. Sci Rep 5:16506. doi:10.1038/srep16506
Manning BD, Snyder M (2000) Drivers and passengers wanted! the role of kinesin-associated proteins. Trends Cell Biol 10(7):281–289
Bansal SK, Gupta N, Sankhwar SN, Rajender S (2015) Differential genes expression between fertile and infertile spermatozoa revealed by transcriptome analysis. PLoS One 10(5):e0127007. doi:10.1371/journal.pone.0127007
Ratan R, Mason DA, Sinnot B, Goldfarb DS, Fleming RJ (2008) Drosophila importin alpha1 performs paralog-specific functions essential for gametogenesis. Genetics 178(2):839–850. doi:10.1534/genetics.107.081778
Jabbour L, Welter JF, Kollar J, Hering TM (2003) Sequence, gene structure, and expression pattern of CTNNBL1, a minor-class intron-containing gene—evidence for a role in apoptosis. Genomics 81(3):292–303
Ganesh K, Adam S, Taylor B, Simpson P, Rada C, Neuberger M (2011) CTNNBL1 is a novel nuclear localization sequence-binding protein that recognizes RNA-splicing factors CDC5L and Prp31. J Biol Chem 286(19):17091–17102. doi:10.1074/jbc.M110.208769
Huang X, Wang G, Wu Y, Du Z (2013) The structure of full-length human CTNNBL1 reveals a distinct member of the armadillo-repeat protein family. Acta Crystallogr D Biol Crystallogr 69(Pt 8):1598–1608. doi:10.1107/S0907444913011360
Hiroi N, Ito T, Yamamoto H, Ochiya T, Jinno S, Okayama H (2002) Mammalian Rcd1 is a novel transcriptional cofactor that mediates retinoic acid-induced cell differentiation. EMBO J 21(19):5235–5244
Ryu JR, Echarri A, Li R, Pendergast AM (2009) Regulation of cell–cell adhesion by Abi/Diaphanous complexes. Mol Cell Biol 29(7):1735–1748. doi:10.1128/MCB.01483-08
Schulze N, Graessl M, Blancke Soares A, Geyer M, Dehmelt L, Nalbant P (2014) FHOD1 regulates stress fiber organization by controlling the dynamics of transverse arcs and dorsal fibers. J Cell Sci 127(Pt 7):1379–1393. doi:10.1242/jcs.134627
Kim HC, Jo YJ, Kim NH, Namgoong S (2015) Small molecule inhibitor of formin homology 2 domains (SMIFH2) reveals the roles of the formin family of proteins in spindle assembly and asymmetric division in mouse oocytes. PLoS One 10(4):e0123438. doi:10.1371/journal.pone.0123438
Xie C, Jiang G, Fan C, Zhang X, Zhang Y, Miao Y, Lin X, Wu J, Wang L, Liu Y, Yu J, Yang L, Zhang D, Xu K, Wang E (2014) ARMC8alpha promotes proliferation and invasion of non-small cell lung cancer cells by activating the canonical Wnt signaling pathway. Tumour Biol 35(9):8903–8911. doi:10.1007/s13277-014-2162-z
Acknowledgments
We thank Dr. Amin Bredan for critical reading and careful editing of the manuscript and our colleagues for helpful discussions. This work was supported by the Research Foundation—Flanders (FWO-Vlaanderen, Award G.0320.11N), the Belgian Science Policy (Interuniversity Attraction Poles—Award IAP7/07), and the Special Research Fund of Ghent University (Award BOF 01J14211).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gul, I.S., Hulpiau, P., Saeys, Y. et al. Metazoan evolution of the armadillo repeat superfamily. Cell. Mol. Life Sci. 74, 525–541 (2017). https://doi.org/10.1007/s00018-016-2319-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-016-2319-6