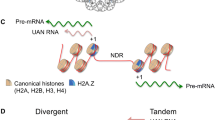
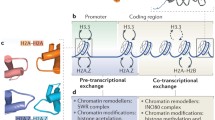
Abstract
Histones are the primary protein component of chromatin, the mixture of DNA and proteins that packages the genetic material in eukaryotes. Large amounts of histones are required during the S phase of the cell cycle when genome replication occurs. However, ectopic expression of histones during other cell cycle phases is toxic; thus, histone expression is restricted to the S phase and is tightly regulated at multiple levels, including transcriptional, post-transcriptional, translational, and post-translational. In this review, we discuss mechanisms of regulation of histone gene expression with emphasis on the transcriptional regulation of the replication-dependent histone genes in the model yeast Saccharomyces cerevisiae.


Similar content being viewed by others
References
Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251–260
Arents G, Burlingame RW, Wang BC, Love WE, Moudrianakis EN (1991) The nucleosomal core histone octamer at 3.1 A resolution: a tripartite protein assembly and a left-handed superhelix. Proc Natl Acad Sci USA 88:10148–10152
Billon P, Cote J (2012) Precise deposition of histone H2A.Z in chromatin for genome expression and maintenance. Biochim Biophys Acta 1819:290–302
Fillingham J, Keogh MC, Krogan NJ (2006) GammaH2AX and its role in DNA double-strand break repair. Biochem Cell Biol 84:568–577
Hereford LM, Osley MA, Ludwig TR 2nd, McLaughlin CS (1981) Cell-cycle regulation of yeast histone mRNA. Cell 24:367–375
Kim UJ, Han M, Kayne P, Grunstein M (1988) Effects of histone H4 depletion on the cell cycle and transcription of Saccharomyces cerevisiae. EMBO J 7:2211–2219
Gunjan A, Verreault A (2003) A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell 115:537–549
Marzluff WF, Gongidi P, Woods KR, Jin J, Maltais LJ (2002) The human and mouse replication-dependent histone genes. Genomics 80:487–498
Smith MM, Murray K (1983) Yeast H3 and H4 histone messenger RNAs are transcribed from two non-allelic gene sets. J Mol Biol 169:641–661
Hereford L, Fahrner K, Woolford J Jr, Rosbash M, Kaback DB (1979) Isolation of yeast histone genes H2A and H2B. Cell 18:1261–1271
Osley MA, Gould J, Kim S, Kane MY, Hereford L (1986) Identification of sequences in a yeast histone promoter involved in periodic transcription. Cell 45:537–544
Prado F, Aguilera A (2005) Partial depletion of histone H4 increases homologous recombination-mediated genetic instability. Mol Cell Biol 25:1526–1536
Han M, Chang M, Kim UJ, Grunstein M (1987) Histone H2B repression causes cell-cycle-specific arrest in yeast: effects on chromosomal segregation, replication, and transcription. Cell 48:589–597
Meeks-Wagner D, Hartwell LH (1986) Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell 44:43–52
Herrero AB, Moreno S (2011) Lsm1 promotes genomic stability by controlling histone mRNA decay. EMBO J 30:2008–2018
Berloco M, Fanti L, Breiling A, Orlando V, Pimpinelli S (2001) The maternal effect gene, abnormal oocyte (abo), of Drosophila melanogaster encodes a specific negative regulator of histones. Proc Natl Acad Sci USA 98:12126–12131
Jong AY, Kuo CL, Campbell JL (1984) The CDC8 gene of yeast encodes thymidylate kinase. J Biol Chem 259:11052–11059
Lycan DE, Osley MA, Hereford LM (1987) Role of transcriptional and posttranscriptional regulation in expression of histone genes in Saccharomyces cerevisiae. Mol Cell Biol 7:614–621
Breeden L (1988) Cell cycle-regulated promoters in budding yeast. Trends Genet 4:249–253
Osley MA, Lycan D (1987) Trans-acting regulatory mutations that alter transcription of Saccharomyces cerevisiae histone genes. Mol Cell Biol 7:4204–4210
Spector MS, Raff A, DeSilva H, Lee K, Osley MA (1997) Hir1p and Hir2p function as transcriptional corepressors to regulate histone gene transcription in the Saccharomyces cerevisiae cell cycle. Mol Cell Biol 17:545–552
Xu H, Kim UJ, Schuster T, Grunstein M (1992) Identification of a new set of cell cycle-regulatory genes that regulate S-phase transcription of histone genes in Saccharomyces cerevisiae. Mol Cell Biol 12:5249–5259
Green EM, Antczak AJ, Bailey AO, Franco AA, Wu KJ, Yates JR 3rd, Kaufman PD (2005) Replication-independent histone deposition by the HIR complex and Asf1. Curr Biol 15:2044–2049
Prochasson P, Florens L, Swanson SK, Washburn MP, Workman JL (2005) The HIR corepressor complex binds to nucleosomes generating a distinct protein/DNA complex resistant to remodeling by SWI/SNF. Genes Dev 19:2534–2539
Fillingham J, Kainth P, Lambert JP, van Bakel H, Tsui K, Pena-Castillo L, Nislow C, Figeys D, Hughes TR, Greenblatt J et al (2009) Two-color cell array screen reveals interdependent roles for histone chaperones and a chromatin boundary regulator in histone gene repression. Mol Cell 35:340–351
Amin AD, Dimova DK, Ferreira ME, Vishnoi N, Hancock LC, Osley MA, Prochasson P (2012) The mitotic Clb cyclins are required to alleviate HIR-mediated repression of the yeast histone genes at the G1/S transition. Biochim Biophys Acta 1819:16–27
Balaji S, Iyer LM, Aravind L (2009) HPC2 and ubinuclein define a novel family of histone chaperones conserved throughout eukaryotes. Mol Biosyst 5:269–275
Nelson DM, Ye X, Hall C, Santos H, Ma T, Kao GD, Yen TJ, Harper JW, Adams PD (2002) Coupling of DNA synthesis and histone synthesis in S phase independent of cyclin/cdk2 activity. Mol Cell Biol 22:7459–7472
Mahajan K, Fang B, Koomen JM, Mahajan NP (2012) H2B Tyr37 phosphorylation suppresses expression of replication-dependent core histone genes. Nat Struct Mol Biol 19:930–937
Banumathy G, Somaiah N, Zhang R, Tang Y, Hoffmann J, Andrake M, Ceulemans H, Schultz D, Marmorstein R, Adams PD (2009) Human UBN1 is an ortholog of yeast Hpc2p and has an essential role in the HIRA/ASF1a chromatin-remodeling pathway in senescent cells. Mol Cell Biol 29:758–770
Rai TS, Puri A, McBryan T, Hoffman J, Tang Y, Pchelintsev NA, van Tuyn J, Marmorstein R, Schultz DC, Adams PD (2011) Human CABIN1 is a functional member of the human HIRA/UBN1/ASF1a histone H3.3 chaperone complex. Mol Cell Biol 31:4107–4118
Pchelintsev NA, McBryan T, Rai TS, van Tuyn J, Ray-Gallet D, Almouzni G, Adams PD (2013) Placing the HIRA histone chaperone complex in the chromatin landscape. Cell Rep 3:1012–1019
Sutton A, Bucaria J, Osley MA, Sternglanz R (2001) Yeast ASF1 protein is required for cell cycle regulation of histone gene transcription. Genetics 158:587–596
Zunder RM, Rine J (2012) Direct interplay among histones, histone chaperones, and a chromatin boundary protein in the control of histone gene expression. Mol Cell Biol 32:4337–4349
Scholes DT, Banerjee M, Bowen B, Curcio MJ (2001) Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics 159:1449–1465
Huang S, Zhou H, Katzmann D, Hochstrasser M, Atanasova E, Zhang Z (2005) Rtt106p is a histone chaperone involved in heterochromatin-mediated silencing. Proc Natl Acad Sci USA 102:13410–13415
Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY (2004) Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22:1567–1572
Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B et al (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387–391
Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H et al (2001) Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294:2364–2368
Tong AH, Drees B, Nardelli G, Bader GD, Brannetti B, Castagnoli L, Evangelista M, Ferracuti S, Nelson B, Paoluzi S et al (2002) A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science 295:321–324
Ferreira ME, Flaherty K, Prochasson P (2011) The Saccharomyces cerevisiae histone chaperone Rtt106 mediates the cell cycle recruitment of SWI/SNF and RSC to the HIR-dependent histone genes. Plos ONE 6:e21113
Ng HH, Robert F, Young RA, Struhl K (2002) Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes Dev 16:806–819
Tackett AJ, Dilworth DJ, Davey MJ, O’Donnell M, Aitchison JD, Rout MP, Chait BT (2005) Proteomic and genomic characterization of chromatin complexes at a boundary. J Cell Biol 169:35–47
Gradolatto A, Smart SK, Byrum S, Blair LP, Rogers RS, Kolar EA, Lavender H, Larson SK, Aitchison JD, Taverna SD et al (2009) A noncanonical bromodomain in the AAA ATPase protein Yta7 directs chromosomal positioning and barrier chromatin activity. Mol Cell Biol 29:4604–4611
Jambunathan N, Martinez AW, Robert EC, Agochukwu NB, Ibos ME, Dugas SL, Donze D (2005) Multiple bromodomain genes are involved in restricting the spread of heterochromatic silencing at the Saccharomyces cerevisiae HMR-tRNA boundary. Genetics 171:913–922
Gradolatto A, Rogers RS, Lavender H, Taverna SD, Allis CD, Aitchison JD, Tackett AJ (2008) Saccharomyces cerevisiae Yta7 regulates histone gene expression. Genetics 179:291–304
Kurat CF, Lambert JP, van Dyk D, Tsui K, van Bakel H, Kaluarachchi S, Friesen H, Kainth P, Nislow C, Figeys D et al (2011) Restriction of histone gene transcription to S phase by phosphorylation of a chromatin boundary protein. Genes Dev 25:2489–2501
Leachman NT, Brellier F, Ferralli J, Chiquet-Ehrismann R, Tucker RP (2010) ATAD2B is a phylogenetically conserved nuclear protein expressed during neuronal differentiation and tumorigenesis. Dev Growth Differ 52:747–755
Raeder MB, Birkeland E, Trovik J, Krakstad C, Shehata S, Schumacher S, Zack TI, Krohn A, Werner HM, Moody SE et al (2013) Integrated genomic analysis of the 8q24 amplification in endometrial cancers identifies ATAD2 as Essential to MYC-dependent cancers. Plos ONE 8:e54873
Fouret R, Laffaire J, Hofman P, Beau-Faller M, Mazieres J, Validire P, Girard P, Camilleri-Broet S, Vaylet F, Leroy-Ladurie F et al (2012) A comparative and integrative approach identifies ATPase family, AAA domain containing 2 as a likely driver of cell proliferation in lung adenocarcinoma. Clin Cancer Res 18:5606–5616
Caron C, Lestrat C, Marsal S, Escoffier E, Curtet S, Virolle V, Barbry P, Debernardi A, Brambilla C, Brambilla E et al (2010) Functional characterization of ATAD2 as a new cancer/testis factor and a predictor of poor prognosis in breast and lung cancers. Oncogene 29:5171–5181
Phillips JE, Corces VG (2009) CTCF: master weaver of the genome. Cell 137:1194–1211
Lambert JP, Fillingham J, Siahbazi M, Greenblatt J, Baetz K, Figeys D (2010) Defining the budding yeast chromatin-associated interactome. Mol Syst Biol 6:448
Silva AC, Xu X, Kim HS, Fillingham J, Kislinger T, Mennella TA, Keogh MC (2012) The replication-independent histone H3–H4 chaperones HIR, ASF1, and RTT106 co-operate to maintain promoter fidelity. J Biol Chem 287:1709–1718
Eriksson PR, Ganguli D, Nagarajavel V, Clark DJ (2012) Regulation of histone gene expression in budding yeast. Genetics 191:7–20
Moran L, Norris D, Osley MA (1990) A yeast H2A–H2B promoter can be regulated by changes in histone gene copy number. Genes Dev 4:752–763
Compagnone-Post PA, Osley MA (1996) Mutations in the SPT4, SPT5, and SPT6 genes alter transcription of a subset of histone genes in Saccharomyces cerevisiae. Genetics 143:1543–1554
Dimova D, Nackerdien Z, Furgeson S, Eguchi S, Osley MA (1999) A role for transcriptional repressors in targeting the yeast Swi/Snf complex. Mol Cell 4:75–83
Xu F, Zhang K, Grunstein M (2005) Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell 121:375–385
Fassler JS, Winston F (1988) Isolation and analysis of a novel class of suppressor of Ty insertion mutations in Saccharomyces cerevisiae. Genetics 118:203–212
Natsoulis G, Dollard C, Winston F, Boeke JD (1991) The products of the SPT10 and SPT21 genes of Saccharomyces cerevisiae increase the amplitude of transcriptional regulation at a large number of unlinked loci. New Biol 3:1249–1259
Hess D, Liu B, Roan NR, Sternglanz R, Winston F (2004) Spt10-dependent transcriptional activation in Saccharomyces cerevisiae requires both the Spt10 acetyltransferase domain and Spt21. Mol Cell Biol 24:135–143
Mendiratta G, Eriksson PR, Shen CH, Clark DJ (2006) The DNA-binding domain of the yeast Spt10p activator includes a zinc finger that is homologous to foamy virus integrase. J Biol Chem 281:7040–7048
Neuwald AF, Landsman D (1997) GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem Sci 22:154–155
Natsoulis G, Winston F, Boeke JD (1994) The SPT10 and SPT21 genes of Saccharomyces cerevisiae. Genetics 136:93–105
Hess D, Winston F (2005) Evidence that Spt10 and Spt21 of Saccharomyces cerevisiae play distinct roles in vivo and functionally interact with MCB-binding factor, SCB-binding factor and Snf1. Genetics 170:87–94
Dollard C, Ricupero-Hovasse SL, Natsoulis G, Boeke JD, Winston F (1994) SPT10 and SPT21 are required for transcription of particular histone genes in Saccharomyces cerevisiae. Mol Cell Biol 14:5223–5228
Chang JS, Winston F (2011) Spt10 and Spt21 are required for transcriptional silencing in Saccharomyces cerevisiae. Eukaryot Cell 10:118–129
Sherwood PW, Osley MA (1991) Histone regulatory (hir) mutations suppress delta insertion alleles in Saccharomyces cerevisiae. Genetics 128:729–738
Cooper K (2006) Rb, whi it’s not just for metazoans anymore. Oncogene 25:5228–5232
Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO (2001) Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409:533–538
Simon I, Barnett J, Hannett N, Harbison CT, Rinaldi NJ, Volkert TL, Wyrick JJ, Zeitlinger J, Gifford DK, Jaakkola TS et al (2001) Serial regulation of transcriptional regulators in the yeast cell cycle. Cell 106:697–708
Macpherson N, Measday V, Moore L, Andrews B (2000) A yeast taf17 mutant requires the Swi6 transcriptional activator for viability and shows defects in cell cycle-regulated transcription. Genetics 154:1561–1576
Eriksson PR, Ganguli D, Clark DJ (2011) Spt10 and Swi4 control the timing of histone H2A/H2B gene activation in budding yeast. Mol Cell Biol 31:557–572
Recht J, Tsubota T, Tanny JC, Diaz RL, Berger JM, Zhang X, Garcia BA, Shabanowitz J, Burlingame AL, Hunt DF et al (2006) Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc Natl Acad Sci USA 103:6988–6993
Han J, Zhou H, Horazdovsky B, Zhang K, Xu RM, Zhang Z (2007) Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science 315:653–655
Driscoll R, Hudson A, Jackson SP (2007) Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science 315:649–652
Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M et al (2007) Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446:806–810
Masumoto H, Hawke D, Kobayashi R, Verreault A (2005) A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature 436:294–298
Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, Verreault A, Zhang Z (2008) Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell 134:244–255
Williams SK, Truong D, Tyler JK (2008) Acetylation in the globular core of histone H3 on lysine-56 promotes chromatin disassembly during transcriptional activation. Proc Natl Acad Sci USA 105:9000–9005
Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP et al (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440:637–643
Selth L, Svejstrup JQ (2007) Vps75, a new yeast member of the NAP histone chaperone family. J Biol Chem 282:12358–12362
Fillingham J, Recht J, Silva AC, Suter B, Emili A, Stagljar I, Krogan NJ, Allis CD, Keogh MC, Greenblatt JF (2008) Chaperone control of the activity and specificity of the histone H3 acetyltransferase Rtt109. Mol Cell Biol 28:4342–4353
Neumann H, Hancock SM, Buning R, Routh A, Chapman L, Somers J, Owen-Hughes T, van Noort J, Rhodes D, Chin JW (2009) A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol Cell 36:153–163
Xie W, Song C, Young NL, Sperling AS, Xu F, Sridharan R, Conway AE, Garcia BA, Plath K, Clark AT et al (2009) Histone h3 lysine 56 acetylation is linked to the core transcriptional network in human embryonic stem cells. Mol Cell 33:417–427
Takayama Y, Takahashi K (2007) Differential regulation of repeated histone genes during the fission yeast cell cycle. Nucleic Acids Res 35:3223–3237
Chen ES, Saitoh S, Yanagida M, Takahashi K (2003) A cell cycle-regulated GATA factor promotes centromeric localization of CENP-A in fission yeast. Mol Cell 11:175–187
Takayama Y, Mamnun YM, Trickey M, Dhut S, Masuda F, Yamano H, Toda T, Saitoh S (2010) Hsk1- and SCF(Pof3)-dependent proteolysis of S. pombe Ams2 ensures histone homeostasis and centromere function. Dev Cell 18:385–396
Trickey M, Fujimitsu K, Yamano H (2013) Anaphase-promoting complex/cyclosome-mediated proteolysis of Ams2 in the G1 phase ensures the coupling of histone gene expression to DNA replication in fission Yeast. J Biol Chemi 288:928–937
Enserink JM, Kolodner RD (2010) An overview of Cdk1-controlled targets and processes. Cell Div 5:11
Meggio F, Pinna LA (2003) One-thousand-and-one substrates of protein kinase CK2? FASEB J 17:349–368
Lombardi LM, Ellahi A, Rine J (2011) Direct regulation of nucleosome density by the conserved AAA-ATPase Yta7. Proc Natl Acad Sci USA 108:E1302–E1311
Ma T, Van Tine BA, Wei Y, Garrett MD, Nelson D, Adams PD, Wang J, Qin J, Chow LT, Harper JW (2000) Cell cycle-regulated phosphorylation of p220(NPAT) by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev 14:2298–2313
Zhao J, Kennedy BK, Lawrence BD, Barbie DA, Matera AG, Fletcher JA, Harlow E (2000) NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev 14:2283–2297
Ye X, Wei Y, Nalepa G, Harper JW (2003) The cyclin E/Cdk2 substrate p220(NPAT) is required for S-phase entry, histone gene expression, and Cajal body maintenance in human somatic cells. Mol Cell Biol 23:8586–8600
Imbeault D, Gamar L, Rufiange A, Paquet E, Nourani A (2008) The Rtt106 histone chaperone is functionally linked to transcription elongation and is involved in the regulation of spurious transcription from cryptic promoters in yeast. J Biol Chem 283:27350–27354
Marzluff WF, Wagner EJ, Duronio RJ (2008) Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet 9:843–854
Davila Lopez M, Samuelsson T (2008) Early evolution of histone mRNA 3′ end processing. RNA 14:1–10
Fahrner K, Yarger J, Hereford L (1980) Yeast histone mRNA is polyadenylated. Nucleic Acids Res 8:5725–5737
Beggs S, James TC, Bond U (2012) The PolyA tail length of yeast histone mRNAs varies during the cell cycle and is influenced by Sen1p and Rrp6p. Nucleic Acids Res 40:2700–2711
Xu HX, Johnson L, Grunstein M (1990) Coding and noncoding sequences at the 3′ end of yeast histone H2B mRNA confer cell cycle regulation. Mol Cell Biol 10:2687–2694
Campbell SG, Li Del Olmo M, Beglan P, Bond U (2002) A sequence element downstream of the yeast HTB1 gene contributes to mRNA 3′ processing and cell cycle regulation. Mol Cell Biol 22:8415–8425
Chowdhury A, Mukhopadhyay J, Tharun S (2007) The decapping activator Lsm1p-7p-Pat1p complex has the intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs. RNA 13:998–1016
Canavan R, Bond U (2007) Deletion of the nuclear exosome component RRP6 leads to continued accumulation of the histone mRNA HTB1 in S-phase of the cell cycle in Saccharomyces cerevisiae. Nucleic Acids Res 35:6268–6279
Mullen TE, Marzluff WF (2008) Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev 22:50–65
Reis CC, Campbell JL (2007) Contribution of Trf4/5 and the nuclear exosome to genome stability through regulation of histone mRNA levels in Saccharomyces cerevisiae. Genetics 175:993–1010
Vasiljeva L, Buratowski S (2006) Nrd1 interacts with the nuclear exosome for 3′ processing of RNA polymerase II transcripts. Mol Cell 21:239–248
Estruch F, Peiro-Chova L, Gomez-Navarro N, Durban J, Hodge C, Del Olmo M, Cole CN (2009) A genetic screen in Saccharomyces cerevisiae identifies new genes that interact with me67–5, a temperature-sensitive allele of the gene encoding the mRNA export receptor. Mol Genet Genomics 281:125–134
Soto M, Iborra S, Quijada L, Folgueira C, Alonso C, Requena JM (2004) Cell-cycle-dependent translation of histone mRNAs is the key control point for regulation of histone biosynthesis in Leishmania infantum. Biochem J 379:617–625
Laffler TG, Carrino J (1987) Cell-cycle-regulated translation of histone mRNA in Physarum plasmodia. J Bacteriol 169:2291–2293
Cakmakci NG, Lerner RS, Wagner EJ, Zheng L, Marzluff WF (2008) SLIP1, a factor required for activation of histone mRNA translation by the stem-loop binding protein. Mol Cell Biol 28:1182–1194
McLaren RS, Caruccio N, Ross J (1997) Human La protein: a stabilizer of histone mRNA. Mol Cell Biol 17:3028–3036
Schenk L, Meinel DM, Strasser K, Gerber AP (2012) La-motif-dependent mRNA association with Slf1 promotes copper detoxification in yeast. RNA 18:449–461
Rother S, Burkert C, Brunger KM, Mayer A, Kieser A, Strasser K (2010) Nucleocytoplasmic shuttling of the La motif-containing protein Sro9 might link its nuclear and cytoplasmic functions. RNA 16:1393–1401
Gunjan A, Paik J, Verreault A (2006) The emergence of regulated histone proteolysis. Curr Opin Genet Dev 16:112–118
Singh RK, Kabbaj MH, Paik J, Gunjan A (2009) Histone levels are regulated by phosphorylation and ubiquitylation-dependent proteolysis. Nat Cell Biol 11:925–933
Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C (2007) A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet 39:1235–1244
Verzijlbergen KF, van Welsem T, Sie D, Lenstra TL, Turner DJ, Holstege FC, Kerkhoven RM, van Leeuwen F (2011) A barcode screen for epigenetic regulators reveals a role for the NuB4/HAT-B histone acetyltransferase complex in histone turnover. Plos Genet 7:e1002284
Kim HJ, Seol JH, Han JW, Youn HD, Cho EJ (2007) Histone chaperones regulate histone exchange during transcription. EMBO J 26:4467–4474
Fazly A, Li Q, Hu Q, Mer G, Horazdovsky B, Zhang Z (2012) Histone chaperone Rtt106 promotes nucleosome formation using (H3-H4)2 tetramers. J Biol Chem 287:10753–10760
Radovani E, Cadorin M, Shams T, El-Rass S, Karsou AR, Kim HS, Kurat CF, Keogh MC, Greenblatt J, Fillingham JS (2013) The carboxyl terminus of Rtt109 functions in chaperone control of histone acetylation. Eukaryot Cell 12:654–664
Zegerman P, Diffley JF (2010) Checkpoint-dependent inhibition of DNA replication initiation by Sld3 and Dbf4 phosphorylation. Nature 467:474–478
Li Z, Vizeacoumar FJ, Bahr S, Li J, Warringer J, Vizeacoumar FS, Min R, Vandersluis B, Bellay J, Devit M et al (2011) Systematic exploration of essential yeast gene function with temperature-sensitive mutants. Nat Biotechnol 29:361–367
Sopko R, Huang D, Preston N, Chua G, Papp B, Kafadar K, Snyder M, Oliver SG, Cyert M, Hughes TR et al (2006) Mapping pathways and phenotypes by systematic gene overexpression. Mol Cell 21:319–330
Acknowledgments
Work on histone gene regulation in the Fillingham and Andrews laboratories is supported by an NSERC Discovery Grant to J.S.F. (grant number: 386646-2010) and a grant from the Canadian Institutes of Health Research (CIHR) to B.J.A. and Tim Hughes (BMB-210972). C.F.K. was supported by an EMBO long-term fellowship.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Kurat, C.F., Recht, J., Radovani, E. et al. Regulation of histone gene transcription in yeast. Cell. Mol. Life Sci. 71, 599–613 (2014). https://doi.org/10.1007/s00018-013-1443-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-013-1443-9