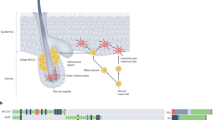
Abstract
Melanoma is a malignant tumor of melanocytes that can spread to other organs of the body, resulting in severe and/or lethal malignancies. Melanocytes are pigment-producing cells found in the deep layer of the epidermis and are originated from melanocytes stem cells through a cellular process called melanogenesis. Several genes and epigenetic and micro-environmental factors are involved in this process via the regulation and maintenance of the balance between melanocytes stem cells proliferation and their differentiation into melanocytes. Dysregulation of this balance through gain or loss of function of key genes implicated in the control and regulation of cell cycle progression and/or differentiation results in melanoma initiation and progression. This review aims to provide a comprehensive overview about the origin of melanocytes, the oncogenic events involved in melanocytes stem cells transformation, and the mechanisms implicated in the perpetuation of melanoma malignant phenotype.





Similar content being viewed by others
References
Erickson CA, Reedy MV (1998) Neural crest development: the interplay between morphogenesis and cell differentiation. Curr Top Dev Biol 40:177–209
Dupin E, Glavieux C, Vaigot P, Le Douarin NM (2000) Endothelin 3 induces the reversion of melanocytes to glia through a neural crest-derived glial-melanocytic progenitor. Proc Natl Acad Sci 97(14):7882–7887
Lahav R, Heffner G, Patterson PH (1999) An endothelin receptor B antagonist inhibits growth and induces cell death in human melanoma cells in vitro and in vivo. Proc Natl Acad Sci 96(20):11496–11500
Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, Iyer V, Jeffrey SS, Van de Rijn M, Waltham M, Pergamenschikov A, Lee JC, Lashkari D, Shalon D, Myers TG, Weinstein JN, Botstein D, Brown PO (2000) Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet 24(3):227–235
Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, Beroukhim R, Milner DA, Granter SR, Du J, Lee C, Wagner SN, Li C, Golub TR, Rimm DL, Meyerson ML, Fisher DE, Sellers WR (2005) Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature 436(7047):117–122
Bonazzi VF, Stark MS, Hayward NK (2012) MicroRNA regulation of melanoma progression. Melanoma Res 22(2):101
Haluska FG, Tsao H, Wu H, Haluska FS, Lazar A, Goel V (2006) Genetic alterations in signaling pathways in melanoma. Clin Cancer Res 12(7):2301s–2307s
Schatton T, Murphy GF, Frank NY, Yamaura K, Waaga-Gasser AM, Gasser M, Zhan Q, Jordan S, Duncan LM, Weishaupt C, Fuhlbrigge RC, Kupper TS, Sayegh MH, Frank MH (2008) Identification of cells initiating human melanomas. Nature 451(7176):345–349
Linley AJ, Mathieu MG, Miles AK, Rees RC, McArdle SE, Regad T (2012) The helicase HAGE expressed by malignant melanoma-initiating cells is required for tumor cell proliferation in vivo. J Biol Chem 287(17):13633–13643
Ikeya M, Lee SM, Johnson JE, McMahon AP, Takada S (1997) Wnt signalling required for expansion of neural crest and CNS progenitors. Nature 389(6654):966–970
Jin EJ, Erickson CA, Takada S, Burrus LW (2001) Wnt and BMP signaling govern lineage segregation of melanocytes in the avian embryo. Dev Biol 233(1):22–37
Dorsky RI, Moon RT, Raible DW (1998) Control of neural crest cell fate by the Wnt signalling pathway. Nature 396(6709):370–373
Dunn KJ, Williams BO, Li Y, Pavan WJ (2000) Neural crest-directed gene transfer demonstrates Wnt1 role in melanocyte expansion and differentiation during mouse development. Proc Natl Acad Sci 97(18):10050–10055
Vance KW, Goding CR (2004) The transcription network regulating melanocyte development and melanoma. Pigment Cell Res 17(4):318–325
Cano A, Pérez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA (2000) The transcription factor snail controls epithelial–mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol 2(2):76–83
Honoré SM, Aybar MJ, Mayor R (2003) Sox10 is required for the early development of the prospective neural crest in Xenopus embryos. Dev Biol 260(1):79–96
Meulemans D, Bronner-Fraser M (2004) Gene-regulatory interactions in neural crest evolution and development. Dev Cell 7(3):291–299
Geissler EN, Ryan MA, Housman DE (1988) The dominant-white spotting (W) locus of the mouse encodes the c-kit proto-oncogene. Cell 55(1):185
Williams DE, Eisenman J, Baird A, Rauch C, Van Ness K, March CJ, Park LS, Martin U, Mochizukl DY, Boswell SH, Burgess GS, Cosman D, Lyman SD (1990) Identification of a ligand for the c-kit proto-oncogene. Cell 63(1):167–174
Wehrle-Haller B, Weston JA (1995) Soluble and cell-bound forms of steel factor activity play distinct roles in melanocyte precursor dispersal and survival on the lateral neural crest migration pathway. Development 121(3):731–742
Parichy DM, Rawls JF, Pratt SJ, Whitfield TT, Johnson SL (1999) Zebrafish sparse corresponds to an orthologue of c-kit and is required for the morphogenesis of a subpopulation of melanocytes, but is not essential for hematopoiesis or primordial germ cell development. Development 126(15):3425–3436
Erickson CA (1993) From the crest to the periphery: control of pigment cell migration and lineage segregation. Pigment Cell Res 6(5):336–347
Nishikawa SI, Osawa M (2007) Generating quiescent stem cells. Pigment Cell Res 20(4):263–270
Fuchs E (2007) Scratching the surface of skin development. Nature 445(7130):834–842
Uong A, Zon LI (2010) Melanocytes in development and cancer. J Cell Physiol 222(1):38–41
Dhomen N, Reis-Filho JS, da Rocha DS, Hayward R, Savage K, Delmas V, Larue L, Pritchard C, Marais R (2009) Oncogenic Braf induces melanocyte senescence and melanoma in mice. Cancer Cell 15(4):294
Daniotti M, Oggionni M, Ranzani T, Vallacchi V, Campi V, Di Stasi D, Torre GD, Perrone F, Luoni C, Suardi S, Frattini M, Pilotti S, Anichini A, Tragni G, Parmiani G, Pierotti MA, Rodolfo M (2004) BRAF alterations are associated with complex mutational profiles in malignant melanoma. Oncogene 23(35):5968–5977
Robinson MJ, Cobb MH (1997) Mitogen-activated protein kinase pathways. Curr Opin Cell Biol 9(2):180–186
Garnett MJ, Marais R (2004) Guilty as charged: B-RAF is a human oncogene. Cancer Cell 6(4):313–319
Thomas NE (2006) BRAF somatic mutations in malignant melanoma and melanocytic naevi. Melanoma Res 16(2):97
Hatzivassiliou G, Song K, Yen I, Brandhuber BJ, Anderson DJ, Alvarado R, Ludlam MJ, Stokoe D, Gloor SL, Vigers G, Morales T, Aliagas I, Liu B, Sideris S, Hoeflich KP, Jaiswal BS, Seshagiri S, Koeppen H, Belvin M, Friedman LS, Malek S (2010) RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature 464(7287):431–435
Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N (2010) RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature 464(7287):427–430
Stark MS, Woods SL, Gartside MG, Bonazzi VF, Dutton-Regester K, Aoude LG, Chow D, Sereduk C, Niemi NM, Tang N, Ellis JJ, Reid J, Zismann V, Tyagi S, Muzny D, Newsham I, Wu Y, Palmer JM, Pollak T, Youngkin D, Brooks BR, Lanagan C, Schmidt CW, Kobe B, MacKeigan JP, Yin H, Brown KM, Gibbs R, Trent J, Hayward NK (2011) Frequent somatic mutations in MAP3K5 and MAP3K9 in metastatic melanoma identified by exome sequencing. Nat Genet 44(2):165–169
Nikolaev SI, Rimoldi D, Iseli C, Valsesia A, Robyr D, Gehrig C, Harshman K, Guipponi M, Bukach O, Zoete V, Michielin O, Muehlethaler K, Speiser D, Beckmann JS, Xenarios I, Halazonetis TD, Jongeneel CV, Stevenson BJ, Antonarakis SE (2011) Exome sequencing identifies recurrent somatic MAP2K1 and MAP2K2 mutations in melanoma. Nat Genet 44(2):133–139. doi:10.1038/ng.1026
Kwong LN, Costello JC, Liu H, Jiang S, Helms TL, Langsdorf AE, Jakubosky D, Genovese G, Muller FL, Jeong JH, Bender RP, Chu GC, Flaherty KT, Wargo JA, Collins JJ, Chin L (2012) Oncogenic NRAS signaling differentially regulates survival and proliferation in melanoma. Nat Med 18(10):1503–1510. doi:10.1038/nm.2941
Houben R, Hesbacher S, Schmid CP, Kauczok CS, Flohr U, Haferkamp S, Müller CS, Schrama D, Wischhusen J, Becker JC (2011) High-level expression of wild-type p53 in melanoma cells is frequently associated with inactivity in p53 reporter gene assays. PLoS One 6(7):e22096
Leibeling D, Laspe P, Emmert S (2006) Nucleotide excision repair and cancer. J Mol Histol 37(5):225–238
De Boer J, Hoeijmakers JH (2000) Nucleotide excision repair and human syndromes. Carcinogenesis 21(3):453–460
Daya-Grosjean L, Sarasin A (2005) The role of UV induced lesions in skin carcinogenesis: an overview of oncogene and tumor suppressor gene modifications in xeroderma pigmentosum skintumors. Mut Res Fundam Mol Mech Mutagen 571(1-2):43–56
Völler D, Ott C, Bosserhoff A (2013) MicroRNAs in malignant melanoma. Clinic Biochem. doi:10.1016/j.clinbiochem.2013.01.008
Mueller DW, Bosserhoff AK (2009) Role of miRNAs in the progression of malignant melanoma. Br J Cancer 101(4):551–556
Felicetti F, Errico MC, Segnalini P, Mattia G, Carè A (2008) MicroRNA-221 and-222 pathway controls melanoma progression. Expert Rev Anticancer Ther 8(11):1759–1765
Penna E, Orso F, Cimino D, Tenaglia E, Lembo A, Quaglino E, Poliseno L, Haimovic A, Osella-Abate S, De Pittà C, Pinatel E, Stadler MB, Provero P, Bernengo MG, Osman I, Taverna D (2011) microRNA-214 contributes to melanoma tumour progression through suppression of TFAP2C. EMBO J 30(10):1990–2007
Boyle GM, Woods SL, Bonazzi VF, Stark MS, Hacker E, Aoude LG, Dutton-Regester K, Cook AL, Sturm RA, Hayward NK (2011) Melanoma cell invasiveness is regulated by miR-211 suppression of the BRN2 transcription factor. Pigment Cell Melanoma Res 24(3):525–537
Levy C, Khaled M, Iliopoulos D, Janas MM, Schubert S, Pinner S, Chen PH, Li S, Fletcher AL, Yokoyama S, Scott KL, Garraway LA, Song JS, Granter SR, Turley SJ, Fisher DE, Novina CD (2010) Intronic miR-211 assumes the tumor suppressive function of its host gene in melanoma. Mol Cell 40(5):841–849
Müller DW, Bosserhoff AK (2008) Integrin β3 expression is regulated by let-7a miRNA in malignant melanoma. Oncogene 27(52):6698–6706
Schultz J, Lorenz P, Gross G, Ibrahim S, Kunz M (2008) MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res 18(5):549–557
Chen J, Feilotter HE, Paré GC, Zhang X, Pemberton JG, Garady C, Lai D, Yang X, Tron VA (2010) MicroRNA-193b represses cell proliferation and regulates cyclin D1 in melanoma. Am J pathol 176(5):2520–2529
Dar AA, Majid S, de Semir D, Nosrati M, Bezrookove V, Kashani-Sabet M (2011) miRNA-205 suppresses melanoma cell proliferation and induces senescence via regulation of E2F1 protein. J Biol Chem 286(19):16606–16614
Cotsarelis G, Sun TT, Lavker RM (1990) Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell 61(7):1329–1337
Nishimura EK, Jordan SA, Oshima H, Yoshida H, Osawa M, Moriyama M, Jackson IJ, Barrandon Y, Miyachi Y, Nishikawa SI (2002) Dominant role of the niche in melanocyte stem-cell fate determination. Nature 416(6883):854–860
Nishimura EK, Granter SR, Fisher DE (2005) Mechanisms of hair graying: incomplete melanocyte stem cell maintenance in the niche. Science 307(5710):720–724
Steingrímsson E, Copeland NG, Jenkins NA (2005) Melanocyte stem cell maintenance and hair graying. Cell 121(1):9–12
Sarin KY, Artandi SE (2007) Aging, graying, and loss of melanocyte stem cells. Stem Cell Rev 3(3):212–217
Moriyama M, Osawa M, Mak SS, Ohtsuka T, Yamamoto N, Han H, Delmas V, Kageyama R, Beermann F, Larue L, Nishikawa SI (2006) Notch signaling via Hes1 transcription factor maintains survival of melanoblasts and melanocyte stem cells. J Cell Biol 173(3):333–339
Cheli Y, Guiliano S, Botton T, Rocchi S, Hofman V, Hofman P, Hofman V, Hofman P, Bahadoran P, Bertolotto C, Ballotti R (2011) Mitf is the key molecular switch between mouse or human melanoma-initiating cells and their differentiated progeny. Oncogene 30(20):2307–2318
Lang D, Lu MM, Huang L, Engleka KA, Zhang M, Chu EY, Lipner S, Skoultchi A, Millar SE, Epstein JA (2005) Pax3 functions at a nodal point in melanocyte stem cell differentiation. Nature 433(7028):884–887
Schepsky A, Bruser K, Gunnarsson GJ, Goodall J, Hallsson JH, Goding CR, Steingrimsson E, Hecht A (2006) The microphthalmia-associated transcription factor Mitf interacts with β-catenin to determine target gene expression. Mol Cell Biol 26(23):8914–8927
Takeda K, Yasumoto KI, Takada R, Takada S, Watanabe KI, Udono T, Saito H, Takahashi K, Shibahara S (2000) Induction of melanocyte-specific microphthalmia-associated transcription factor by Wnt-3a. J Biol Chem 275(19):14013–14016
Mak SS, Moriyama M, Nishioka E, Osawa M, Nishikawa SI (2006) Indispensable role of Bcl2 in the development of the melanocyte stem cell. Dev Biol 291(1):144–153
Vance KW, Goding CR (2004) The transcription network regulating melanocyte development and melanoma. Pigment Cell Res 17(4):318–325
Hoek KS, Goding CR (2010) Cancer stem cells versus phenotype-switching in melanoma. Pigment cell Melanoma Res 23(6):746–759
Haass NK, Smalley KS, Li L, Herlyn M (2005) Adhesion, migration, and communication in melanocytes and melanoma. Pigment Cell Res 18(3):150–159
Lee JT, Herlyn M (2006) Microenvironmental influences in melanoma progression. J Cell Biochem 101(4):862–872
Hsu MY, Meier F, Herlyn M (2002) Melanoma development and progression: a conspiracy between tumor and host. Differentiation 70(9–10):522–536
Gupta PB, Kuperwasser C, Brunet JP, Ramaswamy S, Kuo WL, Gray JW, Naber SP, Weinberg RA (2005) The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nat Genet 37(10):1047–1054
van Kempen LC, Ruiter DJ, van Muijen GN, Coussens LM (2003) The tumor microenvironment: a critical determinant of neoplastic evolution. Eur J Cell Biol 82(11):539–548
Norrby K (2002) Mast cells and angiogenesis. Apmis 110(5):355–371
Yong LCJ (1997) The mast cell: origin, morphology, distribution, and function. Exp Toxicol Pathol 49(6):409–424
Fang KC, Raymond WW, Blount JL, Caughey GH (1997) Dog mast cell α-chymase activates progelatinase B by cleaving the Phe88-Gln89 and Phe91-Glu92 bonds of the catalytic domain. J Biol Chem 272(41):25628–25635
Braeuer RR, Zigler M, Villares GJ, Dobroff AS, Bar-Eli M (2011) Transcriptional control of melanoma metastasis: the importance of the tumor microenvironment. Semin Cancer Biol 21(2):83–88
Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA (2004) Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 117(7):927–939
Fang D, Nguyen TK, Leishear K, Finko R, Kulp AN, Hotz S, Van Belle PA, Xu X, Elder DE, Herlyn M (2005) A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res 65(20):9328–9337
Lin K, Baritaki S, Militello L, Malaponte G, Bevelacqua Y, Bonavida B (2010) The role of B-RAF mutations in melanoma and the induction of EMT via dysregulation of the NF-κB/Snail/RKIP/PTEN circuit. Genes Cancer 1(5):409–420
Weiss MB, Abel EV, Mayberry MM, Basile KJ, Berger AC, Aplin AE (2012) TWIST1 Is an ERK1/2 effector that promotes invasion and regulates MMP-1 expression in human melanoma cells. Cancer Res 72(24):6382–6392
Gupta PB, Kuperwasser C, Brunet JP, Ramaswamy S, Kuo WL, Gray JW, Naber SP, Weinberg RA (2005) The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nat Genet 37(10):1047–1054
Fenouille N, Tichet M, Dufies M, Pottier A, Mogha A, Soo JK, Rocchi S, Tartare-Deckert S (2012) The epithelial-mesenchymal transition (EMT) regulatory factor SLUG (SNAI2) is a downstream target of SPARC and AKT in promoting melanoma cell invasion. PLoS One 7(7):e40378
Frank NY, Margaryan A, Huang Y, Schatton T, Waaga-Gasser AM, Gasser M, Sayegh MH, Sadee W, Frank MH (2005) ABCB5-mediated doxorubicin transport and chemoresistance in human malignant melanoma. Cancer Res 65(10):4320–4333
Monzani E, Facchetti F, Galmozzi E, Corsini E, Benetti A, Cavazzin C, Gritti A, Piccinini A, Porro D, Santinami M, Invernici G, Parati E, Alessandri G, La Porta CA (2007) Melanoma contains CD133 and ABCG2 positive cells with enhanced tumourigenic potential. Eur J Cancer 43(5):935–946
Topczewska JM, Postovit LM, Margaryan NV, Sam A, Hess AR, Wheaton WW, Nickoloff BJ, Topczewski J, Hendrix MJ (2006) Embryonic and tumorigenic pathways converge via Nodal signaling: role in melanoma aggressiveness. Nat Med 12(8):925–932
Grichnik JM, Ali WN, Burch JA, Byers JD, Garcia CA, Clark RE, Shea CR (1996) KIT expression reveals a population of precursor melanocytes in human skin. J Invest Dermatol 106(5):967–971
Gottesman MM, Fojo T, Bates SE (2002) Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer 2(1):48–58
Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ (2005) Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer 5(8):615–625
Fang D, Nguyen TK, Leishear K, Finko R, Kulp AN, Hotz S, Van Belle PA, Xu X, Elder DE, Herlyn M (2005) A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res 65(20):9328–9337
Monzani E, Facchetti F, Galmozzi E, Corsini E, Benetti A, Cavazzin C, Gritti A, Piccinini A, Porro D, Santinami M, Invernici G, Parati E, Alessandri G, La Porta CA (2007) Melanoma contains CD133- and ABCG2-positive cells with enhanced tumourigenic potential. Eur J Cancer 43(5):935–946
Shmelkov SV, Butler JM, Hooper AT, Hormigo A, Kushner J, Milde T, St Clair R, Baljevic M, White I, Jin DK, Chadburn A, Murphy AJ, Valenzuela DM, Gale NW, Thurston G, Yancopoulos GD, D’Angelica M, Kemeny N, Lyden D, Rafii S (2008) CD133 expression is not restricted to stem cells, and both CD133+ and CD133− metastatic colon cancer cells initiate tumors. J Clin Invest 118(6):2111
La Porta C (2009) Cancer stem cells: lessons from melanoma. Stem Cell Rev 5(1):61–65
Akers SN, Odunsi K, Karpf AR (2010) Regulation of cancer germline antigen gene expression: implications for cancer immunotherapy. Future Oncol 6(5):717–770
Martelange V, De Smet C, De Plaen E, Lurquin C, Boon T (2000) Identification on a human sarcoma of two new genes with tumor-specific expression. Cancer Res 60:3848–3855
Caruthers JM, McKay DB (2002) Helicase structure and mechanism. Curr Opin Struct Biol 12:123–133
Silverman E, Edwalds-Gilbert G, Lin RJ (2003) DExD/H-box proteins and their partners: helping RNA helicases unwind. Gene 312:1–16
Fuller-Pace FV (2006) DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res 34:4206–4215
Cordin O, Banroques J, Tanner NK, Linder P (2006) The DEAD-box protein family of RNA helicases. Gene 367:17–37
Linder P (2006) Dead-box proteins. Family affair—active and passive players in RNP-remodeling. Nucleic Acids Res 34:4168–4180
Linder P, Jankowsky E (2011) From unwinding to clamping—the DEAD box RNA helicase family. Nat Rev Mol Cell Biol 12:505–516
Camats M, Guil S, Kokolo M, Bach-Elias M (2008) p68 RNA helicase (DDX5) alters activity of cis- and trans-acting factors of the alternative splicing of H-Ras. PLoS One 3:2926
Causevic M, Hislop RG, Kernohan NM, Carey FA, Kay RA, Steele RJ, Fuller-Pace FV (2001) Overexpression and poly-ubiquitylation of the DEAD-box RNA helicase p68 in colorectal tumours. Oncogene 20:7734–7743
Shim H, Shim E, Lee H, Hahn J, Kang D, Lee YS, Jeoung D (2006) CAGE, a novel cancer/testis antigen gene, promotes cell motility by activation ERK and p38 MAPK and downregulating ROS. Mol Cells 21(3):367
Clark EL, Coulson A, Dalgliesh C, Rajan P, Nicol SM, Fleming S, Heer R, Gaughan L, Leung HY, Elliott DJ, Fuller-Pace FV, Robson CN (2008) The RNA helicase p68 is a novel androgen receptor co-activator involved in splicing and is overexpressed in prostate cancer. Cancer Res 6:7938–7946
Kim Y, Park H, Park D, Lee YS, Choe J, Hahn JH, Lee H, Kim YM, Jeoung D (2010) Cancer/testis antigen CAGE exerts negative regulation on p53 expression through HDAC2 and confers resistance to anti-cancer drugs. J Biol Chem 285:25957–25968
Fuller-Pace FV, Moore HC (2011) RNA helicases p68 and p72: multifunctional proteins with important implications for cancer development. Future Oncol 7:239–251
Por E, Byun HJ, Lee EJ, Lim JH, Jung SY, Park I, Kim YM, Jeoung DI, Lee H (2010) The cancer/testis antigen CAGE with oncogenic potential stimulates cell proliferation by up-regulating cyclins D1 and E in an AP-1- and E2F-dependent manner. J Biol Chem 285:14475–14485
Kim Y, Jeoung D (2008) Role of CAGE, a novel cancer/testis antigen, in various cellular processes, including tumorigenesis, cytolytic T lymphocyte induction, and cell motility. J Microbiol Biotechnol 18:600–610
Liggins AP, Lim SH, Soilleux EJ, Pulford K, Banham AH (2010) A panel of cancer-testis genes exhibiting broad-spectrum expression in haematological malignancies. Cancer Immun 10:8
Mathieu MG, Linley AJ, Reeder SP, Badoual C, Tartour E, Rees RC, McArdle SE (2010) HAGE, a cancer/testis antigen expressed at the protein level in a variety of cancers. Cancer Immun 10:2
Downward J (2003) Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer 3:11–22
Shields JM, Pruitt K, McFall A, Shaub A, Der CJ (2000) Understanding Ras: “it ain’t over’til it’s over”. Trends Cell Biol 10:147–154
Castellano E, Downward J (2010) Role of RAS in the regulation of PI3-kinase. Curr Top Microbiol Immunol 346:143–169
Roman-Gomez J, Jimenez-Velasco A, Agirre X, Castillejo JA, Navarro G, San Jose-Eneriz E, Garate L, Cordeu L, Cervantes F, Prosper F, Heiniger A, Torres A (2007) Epigenetic regulation of human cancer/testis antigen gene, HAGE, in chronic myeloid leukemia. Haematologica 92(2):153–162
Chen Q, Lin J, Yao DM, Qian J, Qian W, Yang J, Chai HY, Ma JC, Deng ZQ, Wang CZ, Li Y (2012) Aberrant hypomethylation of DDX43 promoter in myelodysplastic syndrome. Br J Haematol 158(2):293–296. doi:10.1111/j.1365-2141.2012.09138.x
Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, Hogg D, Lorigan P, Lebbe C, Jouary T, Schadendorf D, Ribas A, O’Day SJ, Sosman JA, Kirkwood JM, Eggermont AM, Dreno B, Nolop K, Li J, Nelson B, Hou J, Lee RJ, Flaherty KT, McArthur GA (2011) Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 364(26):2507–2516
Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, Hamid O, Schuchter L, Cebon J, Ibrahim N, Kudchadkar R, Burris HA 3rd, Falchook G, Algazi A, Lewis K, Long GV, Puzanov I, Lebowitz P, Singh A, Little S, Sun P, Allred A, Ouellet D, Kim KB, Patel K, Weber J (2012) Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med 367(18):1694–1703. doi:10.1056/NEJMoa1210093
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Regad, T. Molecular and cellular pathogenesis of melanoma initiation and progression. Cell. Mol. Life Sci. 70, 4055–4065 (2013). https://doi.org/10.1007/s00018-013-1324-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-013-1324-2