Abstract
Functional telomeres are protected from non-homologous end-joining (NHEJ) and homologous recombination (HR) DNA repair pathways. Replication is a critical period for telomeres because of the requirement for reconstitution of functional protected telomere conformations, a process that involves DNA repair proteins. Using knockdown of DNA-PKcs and Rad51 expression in three different cell lines, we demonstrate the respective involvement of NHEJ and HR in the formation of telomere aberrations induced by the G-quadruplex ligand 360A during or after replication. HR contributed to specific chromatid-type aberrations (telomere losses and doublets) affecting the lagging strand telomeres, whereas DNA-PKcs-dependent NHEJ was responsible for sister telomere fusions as a direct consequence of G-quadruplex formation and/or stabilization induced by 360A on parental telomere G strands. NHEJ and HR activation at telomeres altered mitotic progression in treated cells. In particular, NHEJ-mediated sister telomere fusions were associated with altered metaphase-anaphase transition and anaphase bridges and resulted in cell death during mitosis or early G1. Collectively, these data elucidate specific molecular and cellular mechanisms triggered by telomere targeting by the G-quadruplex ligand 360A, leading to cancer cell death.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Telomeres are DNA–protein complexes that function as end caps on eukaryotic chromosomes. They are required for chromosome stability and limit a cell’s lifespan [1–4]. Human telomeric DNA consists of 3–20 kb of tandem repeats of the hexanucleotide sequence 5′-TTAGGG-3′ and a G-rich single strand extending beyond the duplex that forms a 130–210 base overhang [5, 6]. The shelterin protein complex is made up of six telomeric proteins (TRF1, TRF2, POT1, TIN2, Rap1 and TPP1) that associate with telomeric DNA to generate a T-loop, the proposed structure for telomere termini (reviewed in [7, 8]). The T-loop results from the invasion of the telomere overhang into the duplex region of the telomere [9]. It has been proposed that this structure prevents the telomere from being recognised as a double strand break (DSB) (reviewed in [10]).
Replication and reconstitution of protected telomere structures during S/G2 phases of the cell cycle represent critical steps for the cell [11] involving DNA damage signaling and activation of DNA repair proteins such as DNA-dependent protein kinase catalytic subunit (DNA-PKcs) and Rad51. In addition to its role in activating non-homologous end-joining (NHEJ) [12–14], the catalytic subunit of the DNA-PK is required to refashion the blunt ends of leading strand telomeres after replication [15]. On the other hand, Rad51 is a key protein in homologous recombination involved in locating regions of homology and in DNA strand invasion (reviewed in [16]). It has also been proposed that Rad51 is essential for T-loop formation, allowing the formation of the D-loop structure by invasion of the single-stranded telomeric overhang into the double-stranded telomeric DNA [11].
Telomere G-overhangs can fold into four-stranded DNA structures called G-quadruplexes [17, 18]. Selective stabilization of telomeric G-quadruplexes with specific small ligands has been shown in vitro to inhibit telomerase, the enzyme that specifically elongates telomeres and which is frequently dysregulated in cancer [19–23]. G-quadruplex ligands induce telomere shortening and/or instability, triggering apoptosis and/or replicative senescence in cancer cells [19, 22–24]. This function makes G-quadruplex ligands an attractive candidate for the development of anticancer agents [25–27].
The highly selective G-quadruplex ligand 360A is a 2,6-pyridine-dicarboxamide derivative that induces telomere aberrations and apoptosis in both telomerase-positive and negative cancer cell lines [23, 28–30]. We have previously shown that both ATM and ATR prevent activation of inappropriate DNA repair at 360A-induced dysfunctional telomeres [29, 30]. 360A is thus a very efficient and effective tool for investigating mechanisms of telomere maintenance and their role in cell cycle progression. Here, we report the characterization of specific mechanisms of telomere destabilization induced by 360A and processing by DNA repair complexes. Using DNA-PKcs and Rad51 knockdown in three different cell lines, we have determined the respective involvement of NHEJ and HR DNA repair pathways in the generation of 360A-induced telomere aberrations during and/or after telomere replication and the impact of these mechanisms on cell cycle progression and cell death.
Materials and methods
Cells and treatments
HeLa (cervical cancer cells), HCT116 (colorectal carcinoma) and As3wt2 (SV40-transformed fibroblasts) human cells were stably transfected with Epstein-Barr virus (EBV) vectors expressing small interfering RNA (siRNA) targeting Rad51 (Rad51KD cells) or DNA-PKcs (DNA-PKcsKD cells), or with an EBV vector expressing a previously described, inactive siRNA sequence (CtKD HeLa cells) [31]. The same EBV vectors were also used for Rad51 or DNA-PKcs knockdown in HeLa H2B-GFP cells that were kindly provided by Dr Wahl [32]. Knockdown efficiencies were determined by decreased ARN (electronic supplementary material ESM, Fig. 1), changes in protein expression [33], DNA repair activities [34] and by G2/M arrest in response to irradiation (ESM, Fig. 2). Cells were cultured in DMEM medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen), 1 mM HEPES (Invitrogen), and 2 mM glutamine and antibiotics (penicillin, 100 U/ml, streptomycin, 100 μg/ml; Sigma) and were maintained in a 5% CO2 atmosphere at 37°C. Hygromycin B (125 μg/ml; Invitrogen) was added to the culture media to maintain EBV vectors in cells.
The pyridine derivative 360A has been described previously [23] and was dissolved in dimethyl sulfoxide (DMSO). Cells were cultured with either 5 μM 360A or the corresponding concentration of DMSO as a control. The cell culture media was renewed every 3–4 days.
Telomere fluorescent in situ hybridization (Telo-FISH)
Telo-FISH on metaphase spreads was adapted from a previously published protocol [35], described in detail in [29]. Briefly, metaphase spreads fixed in 4% formaldehyde were first hybridized with a Cy3-PNA [(CCCTAA)3; Applied Biosystems] telomere probe following by a second hybridization with a FITC-pan-centromeric probe (Cambio) designed to detect the centromeric region of all chromosomes. Chromosome preparations were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI, 1 μg/ml) and mounted in Fluoromount-G (Southern Biotech). Slides were observed under a fluorescent microscope (Olympus AX70) and image acquisition was performed using Genus software (Genetix). Chromosome numbers, non-telomeric (chromatid and chromosome breaks) and telomeric aberrations were quantified per metaphase as described previously [30]. The numbers of metaphases analyzed in Telo-FISH experiments are reported in ESM Table 1.
Chromosome orientation-FISH (CO-FISH)
CO-FISH facilitates distinguishing between lagging strand and leading strand telomeres of metaphase chromosomes by respective detection of parental TTAGGG and CCCTAA telomere strands after degradation of neosynthesized DNA strands. CO-FISH has been adapted from a previously published protocol [36] which has been previously described in detail [29]. Briefly, cells were cultured in medium containing BrdU at a final concentration of 10 μM and collected after one cell cycle. Metaphase spreads were prepared as described for Telo-FISH and stained with Hoechst dye 33258 (0.5 μg/ml; Sigma), exposed to UV light prior to digestion with exonuclease III (3 U/μl; Promega) to remove newly synthesized DNA strands. After dehydration, slides were hybridized with Cy-3-PNA (CCCATT)3 and FITC-PNA (TTAGGG)3, telomere probes (Applied Biosystems). After washing, chromosomes were counterstained with DAPI. Metaphase spreads were digitally imaged as described above.
Live video microscopy
Cells were plated on glass coverslips that were mounted in a Ludin Chamber (LIS) or in a 12-well plate for 24 h prior to video microscopy experiments. Live microscopy was carried out using an inverted microscope (Olympus IX81) placed in an incubator chamber (LIS) at 37°C and coupled with a CoolSNAP HQ camera (Princetown Instruments) controlled using MetaMorph software (Universal Imaging). Levels of CO2 (5%), O2 (19%) and humidity (95% relative humidity) were controlled during all experiments by an active gas supply system (the brick; LIS). Fluorescent images were captured for 10–15 fields using a ×20 objective and a 50–100 ms exposure time every 2 min for 16 h. All images were converted into 8-bit files before being assembled with the MetaMorph software. The total and mitotic cell counts analyzed are reported in ESM Table 2.
Statistics analyses
Statistical analyses were performed using Statview software (Abacus Concepts). If not specifically stated in the figure legend, the Student’s t test was used to evaluate the statistical significance of mean values between conditions. In each figure, standard errors of the mean (SEM) and statistical significance levels are noted as follows: ***P < 0.001, **P < 0.01 and *P < 0.05.
Results
Knocking down Rad51 or DNA-PKcs does not sensitize cells to the G-quadruplex ligand 360A
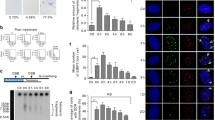
Stable knockdown cell lines have been generated from the human cell lines HeLa (cervical cancer cells), As3wt2 (SV40-transformed fibroblasts) and HCT116 (colorectal carcinoma cells) by expressing small interfering RNA (siRNA) targeted against Rad51 or DNA-PKcs mRNA (referred to as Rad51KD and DNA-PKcsKD cells, respectively). HeLa, As3wt2, and HCT116 cells expressing a nonspecific control siRNA were used as controls (termed CTKD cells; [34]). Although mRNA levels of the siRNA targets were significantly decreased by 60–80% (ESM, Fig. 1a, b), knockdown cell lines exhibited normal growth rates (Fig. 1a) and no increase in chromosome instability (ESM, Fig. 3). Telo-FISH showed that knockdown of Rad51 did not affect telomere stability (Fig. 1e), whereas knockdown of DNA-PKcs resulted in an increase in telomere aberrations in As3wt2 and HCT116 cells (Fig. 1e). These changes consisted mainly of sister telomere losses (***P < 0.001, Fig. 2a) as previously reported in DNA-PKcs−/− HCT116 cells [37].
Rad51 or DNA-PKcs knockdown does not sensitize cells to the G-quadruplex ligand 360A. a Cell growth curves of Rad51KD, DNA-PKcsKD and control (CtKD) HeLa, As3wt2 and HCT116 cells treated for 14 days with 5 μM 360A or 0.05% DMSO as control. b Representative metaphase spread from 360A-treated cells (5 μM for 8 days) obtained by Telo-FISH experiment. Telomeres were hybridized with a telomere Cy3-PNA probe (red) and a centromere DNA probe (green) and chromosomes were counterstained with DAPI (blue). Stars indicate chromosomes with abnormal telomeric signals. Representative examples of non-telomeric aberrations: chromatid (a) or chromosome breaks (b) and chromosome with telomere aberrations: sister telomere fusion (c), telomere doublet (d), sister telomere loss (e), dicentric chromosome (f), terminal deletion (g) and telomeric DNA-containing double minute chromosome (TDM, h). c, d Percentages of chromosomes with chromatid (c) or chromosome breaks (d) induced by 8 days of treatment with the G4-ligand 360A (5 μM) detected in metaphase spreads of Rad51KD, DNA-PKcsKD and control (CtKD) HeLa, As3wt2 and HCT116 cells. All percentages (±SEM) of chromatid and chromosome breaks induced by 360A were calculated from percentages of damaged chromosomes identified in 360A-treated deficient cells minus the mean of damaged chromosomes found in the respective DMSO-treated deficient cells (ESM Fig. 3). The white star indicates a significant difference between DMSO- and 360A-treated cells. Percentages were obtained from 20–40 metaphases per condition (see ESM, Table 1). e Percentage of chromosomes with spontaneous telomeric aberrations per cell as detected by Telo-FISH experiments. f Percentage of telomeric aberrations per cell induced by 8 days of treatment with the G4-ligand 360A (5 μM) as detected by Telo-FISH experiments in metaphase spreads of Rad51KD, DNA-PKcsKD and control (CtKD) HeLa, As3wt2 and HCT116 cells. All percentages (±SEM) of telomeric aberrations induced by 360A were calculated from percentages of chromosomes with damaged telomeres identified in 360A-treated deficient cells minus the mean of chromosomes with damaged telomeres found in the respective DMSO-treated deficient cells (e). White star indicates a significant difference between DMSO- and 360A-treated cells. Percentages were obtained from 20–40 metaphases per condition (see ESM, Table 1)
DNA-PKcs is involved in sister telomere fusions and Rad51 in both sister telomere losses and telomere doublets in 360A-treated cells. a Percentages of chromosomes with the indicated spontaneous telomeric aberrations (sister telomere fusion, telomere doublet and sister telomere loss) identified by Telo-FISH experiments in Rad51KD, DNA-PKcsKD, and control (CtKD) HeLa, As3wt2 and HCT116 cells. b Percentage of chromosomes with the indicated telomeric aberrations per cell induced by 8 days of treatment with the G4-ligand 360A (5 μM) in Rad51KD, DNA-PKcsKD, and control (CtKD) HeLa, As3wt2 or HCT116 cells. All percentages (±SEM) were calculated from the percentages of damaged telomeres found in 360A-treated deficient cells minus the mean of percentage of damaged telomeres found in the respective DMSO-treated deficient cells. A white star indicates a significant difference between DMSO- and 360A-treated cells. Percentages were obtained from 20–40 metaphases per condition (see ESM, Table 1). c Representative chromosome lacking lagging or leading strand telomeres (telomere loss). The right graph presents the respective percentages of single telomere loss affecting the lagging strand (red bars) and the leading strand (green bars) in CtKD, Rad51KD and DNA-PKcsKD HeLa cells treated with 5 μM 360A (plus) or DMSO (minus) for 8 days. Results were obtained from at least 35 telomere losses per condition (Chi-square test, ***P < 0.001). d Representative chromosome with telomere doublets hybridized with the C- probe only (lagging–lagging), with the G- telomeric probe only (leading–leading), or with both probes (lagging–leading) as detected by CO-FISH. The right graph presents the respective percentages of telomere doublets containing two lagging strand telomeres (red bars) or two leading strand telomeres (green bars) or both lagging and leading strand telomeres (speckled red and green bars) in CtKD, Rad51KD and DNA-PKcsKD HeLa cells treated with 5 μM 360A (plus) or DMSO (minus) for 8 days. Results were obtained from at least 60 telomere doublet per condition (Chi-square test, ***P < 0.001)
Cell lines deficient in DNA-PKcs or Rad51 have been shown to be highly sensitive to ionizing radiation. Our DNA-PKcs and Rad51 knockdown cells exhibited an increased G2/M arrest in response to irradiation (ESM, Fig. 2). However, knockdown of either Rad51 or DNA-PKcs did not increase the effects of the G-quadruplex ligand 360A on cell growth and viability in the three cell lines tested (Fig. 1a). On the other hand, treatment of Rad51KD and DNA-PKcsKD cells with 5 μM 360A induced a progressive decline in population doublings and mitotic index, followed by total growth arrest similar to that observed in CTKD cells (Fig. 1a; Table 1).
As previously reported [29, 30], 360A had no effect on the level of chromatid and chromosome breaks in CtKD cells (Fig. 1c, d). Chromosome instability is reported to be increased in DNA-PKcs- and Rad51-deficient cell lines after DNA damage [38, 39]. However, except in Rad51KD As3wt2 cells in which we found a moderate increase in chromatid breaks, 360A did not induce chromatid and chromosome breaks in any of the other DNA-PKcs-and Rad51 knockdown cells (Fig. 1c, d).
We have previously reported that 360A induced a large spectrum of specific telomere damages [29, 30]. Consistent with these reports, Telo-FISH revealed that 5 μM 360A induced the formation of telomeric aberrations in the three CtKD cell lines after 8 days of treatment (***P < 0.001) (Fig. 1b, f). Strikingly, 360A induced either similar (in DNA-PKcsKD HeLa and DNA-PKcsKD As3wt2 cells) or even significantly lower (in the three different Rad51KD cells and in DNA-PKcsKD HCT116 cells) levels of telomere aberrations in knockdown cell lines (Fig. 1f).
Altogether, these data confirm that the G-quadruplex ligand selectively target the telomeres. Moreover, we show that Rad51 or DNA-PKcs knockdown did not sensitize the three cancer cell lines to 360A, and that both Rad51 and DNA-PKcs could be involved in the generation of some telomere aberrations induced by the G-quadruplex ligand.
DNA-PKcs is involved in the formation of sister telomere fusion and Rad51 in sister telomere loss and telomere doublets in 360A-treated cells
Telo-FISH showed that 360A principally induced sister telomere fusions, telomere doublets and sister telomere losses in the three CTKD cells (Figs. 1b and 2b) as we have previously reported for other cell types [29, 30]. Interestingly, the level of induction varied between knockdown cell lines (Fig. 2b), allowing the identification of the respective roles of Rad51 and DNA-PKcs in the generation of these telomere aberrations.
360A induced sister telomere fusions in Rad51KD cell lines as was observed in control cells, but these fusions were not induced in DNA-PKcsKD cells (Fig. 2b). These data clearly indicate that 360A-mediated induction of sister telomere fusions was dependent on DNA-PKcs and not on Rad51, which is consistent with the role of NHEJ in the generation of this type of telomere aberration.
On the other hand, DNA-PKcs knockdown did not influence 360A induction of sister telomere losses and telomere doublets, although some minor variations were observed in different cell contexts. Conversely, 360A induction of sister telomere losses and telomere doublets was significantly decreased or totally inhibited in the Rad51KD cell lines (Fig. 2b). These data provide evidence of a role for Rad51, but not DNA-PKcs, in the induction of these telomere aberrations by 360A.
We have previously reported that sister telomere losses and telomere doublets in 360A-treated cells specifically affected the lagging strand telomeres, which is consistent with the formation/stabilization of G-quadruplexes at the parental G-stands, impairing telomere stability during or after replication [29]. To confirm the involvement of Rad51 in the formation of these telomere aberrations, we performed CO-FISH experiments on metaphase chromosomes from DMSO- and 360A-treated CtKD, Rad51KD and DNA-PKcsKD HeLa cells (Fig. 2c, d). CO-FISH allows for the identification of parental telomeric G and C strands on metaphase chromosomes after degradation of the newly synthesized DNA strands with hybridization with specific probes that are associated with distinct fluorochromes [40]. Equal proportions of chromosomes lacking one signal of lagging or leading strand telomeres were found in untreated knockdown cells as well as in CTKD HeLa cell (Fig. 2c). Consistent with our previous reports, 360A treatment significantly increased the proportion of chromosomes lacking one lagging strand telomere signal in both CtKD and DNA-PKcsKD cells. Similarly, the proportion of telomere doublets made of two lagging strand telomere signals was significantly increased following 360A treatment in both CtKD and DNA-PKcsKD cells (Fig. 2d). In contrast, neither the selective loss of lagging strand telomere signals nor the telomere doublets formed by two lagging strand telomere signals were induced by 360A in Rad51KD HeLa cells, confirming the importance of Rad51 in the generation of these telomere aberrations.
We thus show that the major proteins involved in the HR and NHEJ repair pathways are implicated in the generation of specific telomeric aberrations in 360A-treated cells either during or after telomere replication. The activation of HR as a consequence of the formation and/or stabilization of G-quadruplexes at the parental telomeric G-strands resulted in telomere losses and telomere doublets, whereas activation of NHEJ resulted in sister telomere fusions.
Reduced effects of 360A on mitotic progression in Rad51 and DNA-PKcs knockdowns
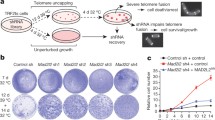
We have previously reported that 360A impaired the progression of mitosis [23]. We therefore investigated the consequences of Rad51 and DNA-PKcs knockdown on mitosis progression. For these experiments, HeLa cells expressing H2B-GFP were transfected with EBV vectors encoding shRNA against DNA-PKcs or Rad51 or an inactive shRNA (ESM, Fig. 1c). Stable clones were then treated with 5 μM 360A and mitosis progression was analyzed by video microscopy after 7 days (ESM, video 1; Fig. 3a).
Approximately 20% of mitoses were found to have an abnormal phenotype in untreated CtKD HeLa cell cultures, which is consistent with the percentages of abnormal mitoses previously reported in HeLa cells (Fig. 3b). This percentage was dramatically increased in 360A-treated CtKD cells, in which more than 45% of mitoses were abnormal (Fig. 3c). Four main types of abnormal phenotypes were induced by 360A in CtKD cells (Fig. 3a): chromosome misalignment at metaphase (ESM, video 2), altered metaphase-anaphase transition characterized by successive phases of chromosome alignment/dealignment (ESM, video 3), anaphase bridges (ESM, video 4), and multipolar mitoses (ESM, video 5). Moreover, 360A induced CTKD cell death either during mitoses (17.8% mitotic cell death; Fig. 3a, g; ESM, video 6) or during early G1 in daughter cells (during the first 3 h after cytokinesis for 15.3% of mitoses; Fig. 3a, g; ESM, video 7) whereas spontaneous cell death was negligible in untreated cells (Fig. 3f).
Telomeric aberrations induced by 360A are involved in mitosis impairment and cell death. a Representative time-lapse images of H2B-GFP HeLa mitosis obtained from ESM, videos 1, 2, 3, 4, 5, 6 and 7, respectively, showing a normal mitosis and examples of abnormal mitoses with chromosome misalignment (white arrow), with an altered metaphase–anaphase transition, with anaphase bridge (white arrow), or multipolar mitosis and cell undergoing a mitotic cell death (white star) or cell death in early G1 of both daughter cells after a normal mitosis (white stars). Original magnification ×20. Elapsed times are noted on each image (min). b Percentage of spontaneous abnormal mitoses in CtKD, Rad51KD and DNA-PKcsKD H2B-GFP HeLa cells. c Percentage of abnormal mitoses induced by 7 days of treatment with 5 μM of the G4-ligand 360A (bottom graph) in CtKD, Rad51KD and DNA-PKcsKD H2B-GFP HeLa cells. All percentages (±SEM) were calculated from the percentages of abnormal mitoses identified in 360A-treated-deficient cells minus the mean of abnormal mitoses found in the respective DMSO-treated deficient cells (b). A white star indicates a significant difference between DMSO- and 360A-treated cells. Percentages were obtained from approximately 200 mitoses per condition (see ESM, Table 2). d Percentage of spontaneous abnormal mitoses with the indicated mitosis abnormality in untreated CtKD, Rad51KD and DNA-PKcsKD H2B-GFP HeLa cells. e Percentage of abnormal mitoses induced by 7 days of treatment with 5 μM of the G4-ligand 360A in CtKD, Rad51KD and DNA-PKcsKD H2B-GFP HeLa cells. All percentages (±SEM) were calculated from percentages of abnormal mitoses found in 360A-treated-deficient cells minus the mean of abnormal mitoses found in the respective DMSO deficient cells. A white star indicates a significant difference between DMSO- and 360A-treated cells. The numbers of analyzed cells are indicated in ESM, Table 2. f Percentage of spontaneous cell death during mitosis or early G1 phase in CtKD, Rad51KD and DNA-PKcsKD HeLa cells treated for 7 days with 0.05% DMSO (open bars) or with 5 μM 360A (closed bars). g Percentage of cell death during mitosis or early G1 phase in CtKD, Rad51KD and DNA-PKcsKD HeLa cells treated for 7 days with 5 μM 360A. All percentages (±SEM) were calculated from the percentages of dead cells found in 360A-treated deficient cells minus the mean of dead cells found in the respective DMSO-treated deficient cells. A white star indicates a significant difference between DMSO- and 360A-treated cells. Numbers of analyzed cells are indicated in ESM, Table 2
Interestingly, whereas the percentages of abnormal phenotypes were similar in untreated knockdown cells and controls (Fig. 3b), percentages of abnormal mitoses induced by 360A were significantly lower in both Rad51KD and DNA-PKcsKD cells than in CtKD HeLa cells (Fig. 3c). This lower induction was not the consequence of selection at mitosis entry, since 360A induced a similar, progressive decrease in mitotic indexes in knockdown and control cells as reported above (Table 2).
Although some variations were detected (Fig. 3d), knockdown of either DNA-PKcs or Rad51 did not significantly impact 360A induction of multipolar mitoses (Fig. 3e). In contrast, 360A induced chromosome misalignments at metaphase in both Rad51KD and DNA-PKcsKD cells (***P < 0.001), but at significantly lower frequencies than in CTKD cells (9.2 and 10% respectively, vs 30.4%, **P < 0.01; Fig. 3e). This observation suggests that both Rad51 and DNA-PKcs could be involved in formation of 360A-induced chromosome misalignments. Furthermore, 360A also induced alterations in metaphase-anaphase transition and anaphase bridges in both CTKD and Rad51KD cells, but these effects were either totally abolished (altered metaphase–anaphase transition) or significantly reduced (anaphase bridges) in DNA-PKcsKD cells (*P < 0.05 and **P < 0.01 respectively; Fig. 3e). Similarly, the induction of cell death during mitosis or in early G1 was significantly reduced in 360A-treated DNA-PKcsKD cells compared to 360A-treated CTKD and -Rad51KD cells (**P < 0.01; Fig. 3g).
Together with our analyses of telomere instability in knockdown cells, these data suggest that DNA-PKcs-mediated sister telomere fusions induced by 360A resulted in anaphase bridges. However, a more extensive study should be conducted to clarify the real implication of DNA-PKcs in more general mitotic problems, as well as the role of Rad51 in mitotic impairment induced by 360A.
Micronuclei induced by 360A are dependent on Rad51
Micronuclei are common indicators of chromosome instability [41, 42]. These small and round DNA/chromatin-containing structures are formed whenever a chromosome or a fragment of chromosome is not incorporated into one of the daughter nuclei or when there is a breakage due to unrepaired or improperly repaired DNA lesions (Fig. 4a). Consistent with a previous report [43], a low number of spontaneous micronuclei was observed in HeLa cells (<2% in untreated CTKD cells; Fig. 4b). Conversely, there was a sevenfold increase in the number of 360A-treated CTKD cells displaying micronuclei (**P < 0.01; 10 μM; Fig. 4b). Telo-FISH and GFP-TRF1 transfection experiments revealed the presence of telomeric sequences and telomeric proteins, respectively, in the majority of 360-induced micronuclei [Fig. 4c, (a) telomeric PNA, red foci and (b) GFP-TRF1, green foci]. In contrast, we did not detect centromeric proteins (after GFP-CENP transfection experiments) or centromeric sequences (PNA centromeric) in 96% of micronuclei. Interestingly, we showed that 360A induced micronuclei in DNA-PKcsKD cells and CtKD cells, but not in Rad51KD cells (Fig. 4b). These data demonstrate that micronuclei formation in 360A-treated cells is Rad51-dependent. Altogether, these data suggest that micronuclei induced by 360A are the direct result of Rad51-dependent telomere aberrations.
G-quadruplex telomere stabilization leads to formation of Rad51-dependant micronuclei containing telomeric sequences. a Representative images of micronuclei induced by 8 days of treatment with 5 μM 360A. Nuclei were counterstained with DAPI. Images a and b are enlargements of the white boxes shown in the left image. Magnification ×60. b Percentages of cells with micronuclei in CtKD, DNA-PKcsKD or Rad51KD HeLa cells cultured with DMSO (open bars) or 360A (5 μM; closed bars) for 8 days. Data were obtained from 300–500 cells per condition. c Micronuclei contain both telomeric sequences (red PNA) and telomeric proteins such as TRF1 (green foci)
Discussion
Both Rad51 and DNA-PKcs have been shown to contribute to telomere protection (reviewed in [44–46]) and it is well established that activation of HR and NHEJ, the two major types of DSB repair pathways, at dysfunctional telomeres can generate telomere aberrations [7, 11, 47–54]. In the present study, we used Rad51 or DNA-PKcs knockdown, resulting in HR or NHEJ deficiencies, respectively, to investigate the telomere destabilization induced by the G-quadruplex ligand 360. We showed that 360A induced both Rad51-dependent telomere aberrations preferentially involving the lagging strand telomeres, including telomere losses or telomere doublets, and DNA-PKcs-dependent sister telomere fusions.
Previous studies have demonstrated that 360A exhibits high selectivity toward quadruplex DNA relative to other DNA structures [23, 28, 55]. Fluorescence titration experiments with oligonucleotides have demonstrated that 360A may induce the formation of a tetramolecular quadruplex, acting as a chaperone for the association of the four strands [56]. Strikingly, 360A induced the G-quadruplex structure more actively than other G-quadruplex ligands tested (telomestatin, TMPyP4 and BRACO19) [56]. Moreover, we have previously demonstrated the specific targeting of telomeres by 360A. Indeed, competitive equilibrium dialysis demonstrated that 360A bound specifically to telomeric-G overhangs from purified genomic DNA, highlighting the selectivity of 360A for telomeric G-quadruplexes [28]. Autoradiography with tritiated-360A evidenced a preferential binding to the terminal region of chromosomes in both human normal and tumor cells [28]. In living cells, 360A induces the degradation of 3′telomeric overhangs and activates a DNA damage signaling in an ATM-dependent manner in cancer cells suggesting that 360A destabilizes telomere structure and induced specific telomere instability [23, 29, 30]. It is noteworthy that neither chromatid nor chromosome breaks were induced by 360A [29, 30] even in DNA-PKcs and Rad51 knockdown cells as shown in this study. Altogether these data emphasize the high specificity of telomere destabilization induced by 360A in various cell lines.
Rad51 is involved in the identification of homologous sequences and strand exchange during HR [57, 58], and may be involved in D-loop formation and establishing the T-loop conformation [11]. The current demonstration of the involvement of HR in both 360A-induced telomere losses and telomere doublets emphasizes our previous hypothesis [29, 30] that the formation/stabilization of G-quadruplexes on parental telomere G overhangs following 360A treatment impairs the reformation of a stable and correct structure at the lagging G-strand, leading to recombination events. Thus, the loss of lagging strand telomeres is likely due to a previously described process, known as T-loop HR, in which unstable T-loops lead to HR-dependent deletion of large segments from individual telomeres [51].
Sfeir et al. [59] reported that conditional deletion of TRF1 in mouse embryonic fibroblasts resulted in multiple telomeric signals, a phenotype that resembles telomeres doublets, as a result of telomere replication problems. Stalled replication forks are processed by HR that catalyzes template switching of blocked replicating strands, which could produce detectable genetic rearrangements when lesions occur in repeated DNA sequences [60]. Such mechanisms could be involved in the generation of telomere doublets as a consequence of stabilized G-quadruplexes, or induced by 360A-dependent impairment of replication fork progression through telomere sequences. This hypothesis is supported by the fact that, contrary to the deletion of TRF1 which produced multiple telomeric signals on both sister telomeres as a consequence of a general problem of telomere replication [59], the telomere doublets in 360A-treated cells involved only the lagging strand telomeres. These findings are consistent with HR activation in order to bypass the presence of G-quadruplex on parental G strands.
We have shown that micronuclei induced by 360A were directly dependent on HR activity. These micronuclei contained telomeric but not centromeric DNA and proteins, suggesting they were not the consequence of a defect in chromosome segregation during mitosis but rather originated from the generation of telomeric DNA fragments dependent on HR activity [61].
NHEJ has been previously shown to be involved in sister telomere fusions [49]. There are several lines of evidence supporting the possible existence of a non-DNA-PKcs-dependent NHEJ pathway [62–65]. It has been proposed that this alternative NHEJ pathway has a predominant role in the formation of chromosome aberrations [65, 66]. However, our data reported here show that this was not the case for 360A-induced sister telomere fusion which involved DNA-PKcs-dependent NHEJ.
DNA-PKcs has been localized to mammalian telomeres [67] and DNA-PKcs−/− mice show increased chromosome end-to-end fusions [68, 69]. Bailey et al. [40] have demonstrated that inhibition of DNA-PKcs exclusively induced chromatid-type fusions involving the leading strand telomeres, indicating that DNA-PKcs is needed for re-establishing a protective terminal structure specifically on telomeres replicated by leading strand DNA synthesis. Persistence of the G-overhang at the lagging telomeres is one of the mechanisms preventing sister telomere fusions prior to the generation of the G-overhang of leading strand telomeres. We propose that G-quadruplexes on the parental telomere G strands lead to resection of the G-overhang at the lagging strand telomeres, which interferes with normal DNA-PKcs activity at leading strand telomeres and subsequently to NHEJ-mediated sister telomere fusions.
One important finding of this study is that Rad51 and DNA-PKcs not only did not protect cells from either telomere instability or the cellular effects induced by 360A but were directly involved in the severe mitotic impairments induced by the G-quadruplex ligand. Liu and colleagues [70] have reported that telomere dysfunction resulting from telomerase deficiency leads to misalignment of metaphase chromosomes during meiotic division in oocytes from late-generation (G4) mice. We have shown that chromosome misalignment at metaphase represents the main type of mitosis abnormality observed in 360A-treated cells. This effect could be directly attributed to the telomere aberrations generated by HR and NHEJ, confirming the necessity of functional telomeres for the correct chromosome alignment at metaphase.
We have demonstrated that DNA-PKcs plays a crucial role in 360A-induced cell death during or just after mitosis. A previous report showed that telomestatin, another G-quadruplex ligand, also induced anaphase bridges in cancer cells [71]. However, the implications of telomere fusions in their formation were not investigated. It would be interesting to investigate the effect of knockdowns of Rad51 and DNA-PKcs under different conditions of telomere destabilization other than 360A. These experiments could delineate the roles of Rad51 and/or DNA-PKcs in the generation of mitotic abnormalities whatever the cause of telomere destabilization.
The lack of both sister telomere fusions and anaphase bridges in 360A-treated DNA-PKcsKD cells clearly demonstrates the involvement of sister telomere fusions in the formation of anaphase bridges induced by the G-quadruplex ligand. Similarly, altered metaphase–anaphase transitions, which were also dependent on DNA-PKcs, likely resulted from a high number of sister telomere fusions delaying or preventing mitotic progression. In the end, DNA breaks resulting from resolution of anaphase bridges and/or the impossibility to progress further through mitosis are probably the cause of cell death during mitosis or early G1, which is thus likely the direct consequence of sister telomere fusion induced by the G-quadruplex ligand.
In summary, we have elucidated some of the molecular and cellular mechanisms triggered by specific telomere targeting by the G-quadruplex ligand 360A, and in particular have determined the roles of HR and NHEJ in generating telomere aberrations leading to cancer cell death.
Abbreviations
- NHEJ:
-
Non-homologous end-joining
- HR:
-
Homologous recombination
- DMSO:
-
Dimethyl sulfoxide
- Telo-FISH:
-
Telomere-fluorescent in situ hybridization
- CO-FISH:
-
Chromosome orientation-FISH
References
Allsopp RC, Vaziri H, Patterson C, Goldstein S, Younglai EV, Futcher AB, Greider CW, Harley CB (1992) Telomere length predicts replicative capacity of human fibroblasts. Proc Natl Acad Sci USA 89(21):10114–10118
Vaziri H, Dragowska W, Allsopp RC, Thomas TE, Harley CB, Lansdorp PM (1994) Evidence for a mitotic clock in human hematopoietic stem cells: loss of telomeric DNA with age. Proc Natl Acad Sci USA 91(21):9857–9860
Blackburn EH (2001) Switching and signaling at the telomere. Cell 106(6):661–673
O’Sullivan RJ, Kubicek S, Schreiber SL, Karlseder J (2010) Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat Struct Mol Biol 17(10):1218–1225
Makarov VL, Hirose Y, Langmore JP (1997) Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell 88(5):657–666
Wright WE, Tesmer VM, Huffman KE, Levene SD, Shay JW (1997) Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev 11(21):2801–2809
de Lange T (2005) Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev 19(18):2100–2110
Palm W, de Lange T (2008) How shelterin protects mammalian telomeres. Annu Rev Genet 42:301–334
Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T (1999) Mammalian telomeres end in a large duplex loop. Cell 97(4):503–514
de Lange T (2002) Protection of mammalian telomeres. Oncogene 21(4):532–540
Verdun RE, Karlseder J (2006) The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell 127(4):709–720
Smith GC, Jackson SP (1999) The DNA-dependent protein kinase. Genes Dev 13(8):916–934
Pastwa E, Blasiak J (2003) Non-homologous DNA end joining. Acta Biochim Pol 50(4):891–908
Burma S, Chen DJ (2004) Role of DNA-PK in the cellular response to DNA double-strand breaks. DNA Repair (Amst) 3(8–9):909–918
Bailey SM, Cornforth MN, Kurimasa A, Chen DJ, Goodwin EH (2001) Strand-specific postreplicative processing of mammalian telomeres. Science 293(5539):2462–2465
Li X, Heyer WD (2008) Homologous recombination in DNA repair and DNA damage tolerance. Cell Res 18(1):99–113
Parkinson GN, Lee MP, Neidle S (2002) Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 417(6891):876–880
Lane AN, Chaires JB, Gray RD, Trent JO (2008) Stability and kinetics of G-quadruplex structures. Nucl Acids Res 36(17):5482–5515
Gowan SM, Harrison JR, Patterson L, Valenti M, Read MA, Neidle S, Kelland LR (2002) A G-quadruplex-interactive potent small-molecule inhibitor of telomerase exhibiting in vitro and in vivo antitumor activity. Mol Pharmacol 61(5):1154–1162
Shammas MA, Shmookler Reis RJ, Akiyama M, Koley H, Chauhan D, Hideshima T, Goyal RK, Hurley LH, Anderson KC, Munshi NC (2003) Telomerase inhibition and cell growth arrest by G-quadruplex interactive agent in multiple myeloma. Mol Cancer Ther 2(9):825–833
Tauchi T, Shin-Ya K, Sashida G, Sumi M, Nakajima A, Shimamoto T, Ohyashiki JH, Ohyashiki K (2003) Activity of a novel G-quadruplex-interactive telomerase inhibitor, telomestatin (SOT-095), against human leukemia cells: involvement of ATM-dependent DNA damage response pathways. Oncogene 22(34):5338–5347
Burger AM, Dai F, Schultes CM, Reszka AP, Moore MJ, Double JA, Neidle S (2005) The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function. Cancer Res 65(4):1489–1496
Pennarun G, Granotier C, Gauthier LR, Gomez D, Hoffschir F, Mandine E, Riou JF, Mergny JL, Mailliet P, Boussin FD (2005) Apoptosis related to telomere instability and cell cycle alterations in human glioma cells treated by new highly selective G-quadruplex ligands. Oncogene 24(18):2917–2928
Zahler AM, Williamson JR, Cech TR, Prescott DM (1991) Inhibition of telomerase by G-quartet DNA structures. Nature 350(6320):718–720
Mergny JL, Helene C (1998) G-quadruplex DNA: a target for drug design. Nat Med 4(12):1366–1367
Han H, Hurley LH (2000) G-quadruplex DNA: a potential target for anti-cancer drug design. Trends Pharmacol Sci 21(4):136–142
Chen B, Liang J, Tian X, Liu X (2008) G-quadruplex structure: a target for anticancer therapy and a probe for detection of potassium. Biochemistry (Mosc) 73(8):853–861
Granotier C, Pennarun G, Riou L, Hoffschir F, Gauthier LR, De Cian A, Gomez D, Mandine E, Riou JF, Mergny JL, Mailliet P, Dutrillaux B, Boussin FD (2005) Preferential binding of a G-quadruplex ligand to human chromosome ends. Nucl Acids Res 33(13):4182–4190
Pennarun G, Granotier C, Hoffschir F, Mandine E, Biard D, Gauthier LR, Boussin FD (2008) Role of ATM in the telomere response to the G-quadruplex ligand 360A. Nucl Acids Res 36(5):1741–1754
Pennarun G, Hoffschir F, Revaud D, Granotier C, Gauthier LR, Mailliet P, Biard DS, Boussin FD (2010) ATR contributes to telomere maintenance in human cells. Nucl Acids Res 38(9):2955–2963
Biard DS, Despras E, Sarasin A, Angulo JF (2005) Development of new EBV-based vectors for stable expression of small interfering RNA to mimick human syndromes: application to NER gene silencing. Mol Cancer Res 3(9):519–529
Kanda T, Sullivan KF, Wahl GM (1998) Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr Biol 8(7):377–385
Despras E, Pfeiffer P, Salles B, Calsou P, Kuhfittig-Kulle S, Angulo JF, Biard DS (2007) Long-term XPC silencing reduces DNA double-strand break repair. Cancer Res 67(6):2526–2534
Biard DS (2007) Untangling the relationships between DNA repair pathways by silencing more than 20 DNA repair genes in human stable clones. Nucl Acids Res 35(11):3535–3550
Lansdorp PM, Verwoerd NP, van de Rijke FM, Dragowska V, Little MT, Dirks RW, Raap AK, Tanke HJ (1996) Heterogeneity in telomere length of human chromosomes. Hum Mol Genet 5(5):685–691
Goodwin E, Meyne J (1993) Strand-specific FISH reveals orientation of chromosome 18 alphoid DNA. Cytogenet Cell Genet 63(2):126–127
Ruis BL, Fattah KR, Hendrickson EA (2008) The catalytic subunit of DNA-dependent protein kinase regulates proliferation, telomere length, and genomic stability in human somatic cells. Mol Cell Biol 28(20):6182–6195
Thacker J (2005) The RAD51 gene family, genetic instability and cancer. Cancer Lett 219(2):125–135
Mahaney BL, Meek K, Lees-Miller SP (2009) Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem J 417(3):639–650
Bailey SM, Goodwin EH, Cornforth MN (2004) Strand-specific fluorescence in situ hybridization: the CO-FISH family. Cytogenet Genome Res 107(1–2):14–17
Iarmarcovai G, Bonassi S, Botta A, Baan RA, Orsiere T (2008) Genetic polymorphisms and micronucleus formation: a review of the literature. Mutat Res 658(3):215–233
Rao X, Zhang Y, Yi Q, Hou H, Xu B, Chu L, Huang Y, Zhang W, Fenech M, Shi Q (2008) Multiple origins of spontaneously arising micronuclei in HeLa cells: direct evidence from long-term live cell imaging. Mutat Res 646(1–2):41–49
Hoffelder DR, Luo L, Burke NA, Watkins SC, Gollin SM, Saunders WS (2004) Resolution of anaphase bridges in cancer cells. Chromosoma 112(8):389–397
Slijepcevic P (2006) The role of DNA damage response proteins at telomeres—an “integrative” model. DNA Repair (Amst) 5(11):1299–1306
Verdun RE, Karlseder J (2007) Replication and protection of telomeres. Nature 447(7147):924–931
Riha K, Heacock ML, Shippen DE (2006) The role of the nonhomologous end-joining DNA double-strand break repair pathway in telomere biology. Annu Rev Genet 40:237–277
van Steensel B, Smogorzewska A, de Lange T (1998) TRF2 protects human telomeres from end-to-end fusions. Cell 92(3):401–413
Karlseder J, Smogorzewska A, de Lange T (2002) Senescence induced by altered telomere state, not telomere loss. Science 295(5564):2446–2449
Smogorzewska A, Karlseder J, Holtgreve-Grez H, Jauch A, de Lange T (2002) DNA ligase IV-dependent NHEJ of deprotected mammalian telomeres in G1 and G2. Curr Biol 12(19):1635–1644
Zhu XD, Niedernhofer L, Kuster B, Mann M, Hoeijmakers JH, de Lange T (2003) ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol Cell 12(6):1489–1498
Wang RC, Smogorzewska A, de Lange T (2004) Homologous recombination generates T-loop-sized deletions at human telomeres. Cell 119(3):355–368
Wu L, Multani AS, He H, Cosme-Blanco W, Deng Y, Deng JM, Bachilo O, Pathak S, Tahara H, Bailey SM, Behringer RR, Chang S (2006) Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell 126(1):49–62
Hsiao SJ, Smith S (2009) Sister telomeres rendered dysfunctional by persistent cohesion are fused by NHEJ. J Cell Biol 184(4):515–526
Williams ES, Klingler R, Ponnaiya B, Hardt T, Schrock E, Lees-Miller SP, Meek K, Ullrich RL, Bailey SM (2009) Telomere dysfunction and DNA-PKcs deficiency: characterization and consequence. Cancer Res 69(5):2100–2107
Lemarteleur T, Gomez D, Paterski R, Mandine E, Mailliet P, Riou JF (2004) Stabilization of the c-myc gene promoter quadruplex by specific ligands’ inhibitors of telomerase. Biochem Biophys Res Commun 323(3):802–808. doi:10.1016/j.bbrc.2004.08.150
De Cian A, Mergny JL (2007) Quadruplex ligands may act as molecular chaperones for tetramolecular quadruplex formation. Nucl Acids Res 35(8):2483–2493. doi:10.1093/nar/gkm098
Moens PB, Freire R, Tarsounas M, Spyropoulos B, Jackson SP (2000) Expression and nuclear localization of BLM, a chromosome stability protein mutated in Bloom’s syndrome, suggest a role in recombination during meiotic prophase. J Cell Sci 113(Pt 4):663–672
Krejci L, Chen L, Van Komen S, Sung P, Tomkinson A (2003) Mending the break: two DNA double-strand break repair machines in eukaryotes. Prog Nucl Acid Res Mol Biol 74:159–201
Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T (2009) Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138(1):90–103
Allen C, Ashley AK, Hromas R, Nickoloff JA (2011) More forks on the road to replication stress recovery. J Mol Cell Biol 3(1):4–12
Norppa H, Falck GC (2003) What do human micronuclei contain? Mutagenesis 18(3):221–233
DiBiase SJ, Zeng ZC, Chen R, Hyslop T, Curran WJ Jr, Iliakis G (2000) DNA-dependent protein kinase stimulates an independently active, nonhomologous, end-joining apparatus. Cancer Res 60(5):1245–1253
Zhu C, Mills KD, Ferguson DO, Lee C, Manis J, Fleming J, Gao Y, Morton CC, Alt FW (2002) Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell 109(7):811–821
Virsik-Kopp P, Rave-Frank M, Hofman-Huther H, Schmidberger H (2003) Role of DNA-PK in the process of aberration formation as studied in irradiated human glioblastoma cell lines M059K and M059J. Int J Radiat Biol 79(1):61–68
Iliakis G, Wang H, Perrault AR, Boecker W, Rosidi B, Windhofer F, Wu W, Guan J, Terzoudi G, Pantelias G (2004) Mechanisms of DNA double strand break repair and chromosome aberration formation. Cytogenet Genome Res 104(1–4):14–20
Revaud D, Martins LM, Boussin FD, Sabatier L, Desmaze C (2011) Different DNA-PKcs functions in the repair of radiation-induced and spontaneous DSBs within interstitial telomeric sequences. Chromosoma 120(3):309–319. doi:10.1007/s00412-011-0313-1
d’Adda di Fagagna F, Hande MP, Tong WM, Roth D, Lansdorp PM, Wang ZQ, Jackson SP (2001) Effects of DNA nonhomologous end-joining factors on telomere length and chromosomal stability in mammalian cells. Curr Biol 11(15):1192–1196
Goytisolo FA, Samper E, Edmonson S, Taccioli GE, Blasco MA (2001) The absence of the DNA-dependent protein kinase catalytic subunit in mice results in anaphase bridges and in increased telomeric fusions with normal telomere length and G-strand overhang. Mol Cell Biol 21(11):3642–3651
Gilley D, Tanaka H, Hande MP, Kurimasa A, Li GC, Oshimura M, Chen DJ (2001) DNA-PKcs is critical for telomere capping. Proc Natl Acad Sci USA 98(26):15084–15088
Liu L, Blasco MA, Keefe DL (2002) Requirement of functional telomeres for metaphase chromosome alignments and integrity of meiotic spindles. EMBO Rep 3(3):230–234
Tahara H, Shin-Ya K, Seimiya H, Yamada H, Tsuruo T, Ide T (2006) G-Quadruplex stabilization by telomestatin induces TRF2 protein dissociation from telomeres and anaphase bridge formation accompanied by loss of the 3′ telomeric overhang in cancer cells. Oncogene 25(13):1955–1966
Acknowledgments
The authors thank Dr Geoffrey M. Wahl (The Salk Institute for Biological Studies, La Jolla, CA, USA) for the HeLa H2B-GFP cell line and Dr Jan Karlseder (MCBL, SIBS, La Jolla, CA, USA) for providing advice and helpful discussion.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material. All videos have an original magnification of ×20 and a time exposure of 75 ms.
18_2011_767_MOESM5_ESM.mov
Time-lapse video microscopy showing chromosome misalignment during mitosis in a H2B-GFP HeLa cell after 7 days of culture with 5 μM 360A. (MOV 825 kb)
18_2011_767_MOESM6_ESM.mov
Time-lapse video microscopy of mitosis with altered metaphase-anaphase transition in a H2B-GFP HeLa cell after 7 days of culture with 5 μM 360A (MOV 811 kb)
18_2011_767_MOESM7_ESM.mov
Time-lapse video microscopy showing an anaphase bridge during mitosis in a H2B-GFP HeLa cell after 7 days of culture with 5 μM 360A (MOV 382 kb)
18_2011_767_MOESM8_ESM.mov
Time-lapse video microscopy of a multipolar mitosis in a H2B-GFP HeLa cell after 7 days of culture with 5 μM 360A (MOV 774 kb)
18_2011_767_MOESM9_ESM.mov
Time-lapse video microscopy of a H2B-GFP HeLa cell undergoing mitotic cell death after 7 days of culture with 5 μM 360A (MOV 550 kb)
18_2011_767_MOESM10_ESM.mov
Time-lapse video microscopy of a normal mitosis in a 360A-treated H2B-GFP HeLa cell, followed by cell death in early G1 of daughter cells 90 min after the abscission step of cytokinesis 10 (MOV 1129 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Gauthier, L.R., Granotier, C., Hoffschir, F. et al. Rad51 and DNA-PKcs are involved in the generation of specific telomere aberrations induced by the quadruplex ligand 360A that impair mitotic cell progression and lead to cell death. Cell. Mol. Life Sci. 69, 629–640 (2012). https://doi.org/10.1007/s00018-011-0767-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-011-0767-6