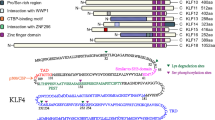
Abstract
Krueppel-like factor 4 (Klf4) belongs to the Sp/Klf family of zinc-finger transcription factors and is indispensable for terminal maturation of epithelial tissues. Furthermore, it is part of a small set of proteins that are used to generate pluripotent embryonic stem cells from differentiated tissues. Herein, we describe that a Klf4 zinc-finger domain mutant induces self-renewal and block of maturation, while wild-type Klf4 induces terminal macrophage differentiation. Moreover, we present the crystal structure of the zinc-finger domain of Klf4 bound to its target DNA, revealing that primarily the two C-terminal zinc-finger motifs are required for site specificity. Lack of those two zinc fingers leads to deficiency of Klf4 to induce macrophage differentiation. The first zinc finger, on the other hand, inhibits the otherwise cryptic self-renewal and block of differentiation activity of Klf4. Our data show that impairing the DNA binding could potentially contribute to a monocytic leukemia.




Similar content being viewed by others
Abbreviations
- Klf4:
-
Krueppel-like factor 4
References
Shields JM, Yang VW (1998) Identification of the DNA sequence that interacts with the gut-enriched Krüppel-like factor. Nucleic Acids Res 26:796–802
Pearson R, Fleetwood J, Eaton S, Crossley M, Bao S (2008) Krüppel-like transcription factors: a functional family. Int J Biochem Cell Biol 40:1996–2001
Suske G, Bruford E, Philipsen S (2005) Mammalian SP/KLF transcription factors: bring in the family. Genomics 85:551–556
Safe S, Abdelrahim M (2005) Sp transcription factor family and its role in cancer. Eur J Cancer 41:2438–2448
Kaczynski J, Cook T, Urrutia R (2003) Sp1- and Krüppel-like transcription factors. Genome Biol 4:206
Nuez B, Michalovich D, Bygrave A, Ploemacher R, Grosveld F (1995) Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature 375:316–318
Perkins AC, Sharpe AH, Orkin SH (1995) Lethal beta-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature 375:318–322
Kuo CT, Veselits ML, Leiden JM (1997) LKLF: a transcriptional regulator of single-positive T cell quiescence and survival. Science 277:1986–1990
Carlson CM, Endrizzi BT, Wu J, Ding X, Weinreich MA, Walsh ER, Wani MA, Lingrel JB, Hogquist KA, Jameson SC (2006) Krüppel-like factor 2 regulates thymocyte and T-cell migration. Nature 442:299–302
Alder JK, Georgantas RW 3rd, Hildreth RL, Kaplan IM, Morisot S, Yu X, McDevitt M, Civin CI (2008) Krüppel-like factor 4 is essential for inflammatory monocyte differentiation in vivo. J Immunol 180:5645–5652
Feinberg MW, Wara AK, Cao Z, Lebedeva MA, Rosenbauer F, Iwasaki H, Hirai H, Katz JP, Haspel RL, Gray S, Akashi K, Segre J, Kaestner KH, Tenen DG, Jain MK (2007) The Krüppel-like factor KLF4 is a critical regulator of monocyte differentiation. EMBO J 26:4138–4148
Zaehres H, Scholer HR (2007) Induction of pluripotency: from mouse to human. Cell 131:834–835
Yet SF, McA’Nulty MM, Folta SC, Yen HW, Yoshizumi M, Hsieh CM, Layne MD, Chin MT, Wang H, Perrella MA, Jain MK, Lee ME (1998) Human EZF, a Krüppel-like zinc finger protein, is expressed in vascular endothelial cells and contains transcriptional activation and repression domains. J Biol Chem 273:1026–1031
Pavletich NP, Pabo CO (1991) Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 Å. Science 252:809–817
Stoll R, Lee BM, Debler EW, Laity JH, Wilson IA, Dyson HJ, Wright PE (2007) Structure of the Wilms tumor suppressor protein zinc finger domain bound to DNA. J Mol Biol 372:1227–1245
Pavletich NP, Pabo CO (1993) Crystal structure of a five-finger GLI-DNA complex: new perspectives on zinc fingers. Science 261:1701–1707
Nolte RT, Conlin RM, Harrison SC, Brown RS (1998) Differing roles for zinc fingers in DNA recognition: structure of a six-finger transcription factor IIIA complex. Proc Natl Acad Sci USA 95:2938–2943
Oka S, Shiraishi Y, Yoshida T, Ohkubo T, Sugiura Y, Kobayashi Y (2004) NMR structure of transcription factor Sp1 DNA binding domain. Biochemistry 43:16027–16035
Simpson RJ, Cram ED, Czolij R, Matthews JM, Crossley M, Mackay JP (2003) CCHX zinc finger derivatives retain the ability to bind Zn(II) and mediate protein–DNA interactions. J Biol Chem 278:28011–28018
Scheich C, Kümmel D, Soumailakakis D, Heinemann U, Büssow K (2007) Vectors for co-expression of an unrestricted number of proteins. Nucleic Acids Res 35:e43
Heinemann U, Büssow K, Mueller U, Umbach P (2003) Facilities and methods for the high-throughput crystal structural analysis of human proteins. Acc Chem Res 36:157–163
Schneider TR, Sheldrick GM (2002) Substructure solution with SHELXD. Acta Crystallogr D 58:1772–1779
Vonrhein C, Blanc E, Roversi P, Bricogne G (2007) Automated structure solution with autoSHARP. Methods Mol Biol 364:215–230
Perrakis A, Morris R, Lamzin VS (1999) Automated protein model building combined with iterative structure refinement. Nat Struct Biol 6:458–463
Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D 60:2126–2132
Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D 53:240–255
McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ (2005) Likelihood-enhanced fast translation functions. Acta Crystallogr D 61:458–464
DeLano WL (2010) The PyMOL molecular graphics system. DeLano Scientific, San Carlos. http://www.pymol.org
Feil R, Wagner J, Metzger D, Chambon P (1997) Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun 237:752–757
Krissinel E, Henrick K (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D 60:2256–2268
Elrod-Erickson M, Rould MA, Nekludova L, Pabo CO (1996) Zif268 protein-DNA complex refined at 1.6 Å: a model system for understanding zinc finger–DNA interactions. Structure 4:1171–1180
Houbaviy HB, Usheva A, Shenk T, Burley SK (1996) Cocrystal structure of YY1 bound to the adeno-associated virus P5 initiator. Proc Natl Acad Sci USA 93:13577–13582
Siatecka M, Sahr KE, Andersen SG, Mezei M, Bieker JJ, Peters LL (2010) Severe anemia in the Nan mutant mouse caused by sequence-selective disruption of erythroid Kruppel-like factor. Proc Natl Acad Sci USA 107:15151–15156
Wolfe SA, Nekludova L, Pabo CO (2000) DNA recognition by Cys2His2 zinc finger proteins. Annu Rev Biophys Biomol Struct 29:183–212
Lavery R, Sklenar H (1988) The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J Biomol Struct Dyn 6:63–91
Heinemann U, Alings C, Hahn M (1994) Crystallographic studies of DNA helix structure. Biophys Chem 50:157–167
Heinemann U, Alings C (1989) Crystallographic study of one turn of G/C-rich B-DNA. J Mol Biol 210:369–381
Heinemann U, Alings C (1991) The conformation of a B-DNA decamer is mainly determined by its sequence and not by crystal environment. EMBO J 10:35–43
Tolstorukov MY, Colasanti AV, McCandlish DM, Olson WK, Zhurkin VB (2007) A novel roll-and-slide mechanism of DNA folding in chromatin: implications for nucleosome positioning. J Mol Biol 371:725–738
Littlewood TD, Hancock DC, Danielian PS, Parker MG, Evan GI (1995) A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res 23:1686–1690
Shields JM, Christy RJ, Yang VW (1996) Identification and characterization of a gene encoding a gut-enriched Krüppel-like factor expressed during growth arrest. J Biol Chem 271:20009–20017
Schaller E, Macfarlane AJ, Rupec RA, Gordon S, McKnight AJ, Pfeffer K (2002) Inactivation of the F4/80 glycoprotein in the mouse germ line. Mol Cell Biol 22:8035–8043
Geiman DE, Ton-That H, Johnson JM, Yang VW (2000) Transactivation and growth suppression by the gut-enriched Krüppel-like factor (Krüppel-like factor 4) are dependent on acidic amino acid residues and protein–protein interaction. Nucleic Acids Res 28:1106–1113
Wei Z, Yang Y, Zhang P, Andrianakos R, Hasegawa K, Lyu J, Chen X, Bai G, Liu C, Pera M, Lu W (2009) Klf4 interacts directly with Oct4 and Sox2 to promote reprogramming. Stem Cells 27:2969–2978
Vardiman JW, Harris NL, Brunning RD (2002) The World Health Organization (WHO) classification of the myeloid neoplasms. Blood 100:2292–2302
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft to D.C. (CA 306/1-1; 1-2) and the “Deutsche Krebshilfe” to D.C. The Protein Sample Production Facility at the Max Delbrück Center is funded by the Helmholtz Association of German Research Centres. We thank Janett Tischer, Silke Kurths, Ingrid Berger, and Tracy Dornblut for excellent technical assistance. We are also thankful to the kind staff of the cell sorting facility of the Deutsche Rheumaforschungszentrum and acknowledge Uwe Müller and the beamline support from the staff of Helmholtz-Zentrum Berlin für Materialien und Energie at BESSY.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Schuetz, A., Nana, D., Rose, C. et al. The structure of the Klf4 DNA-binding domain links to self-renewal and macrophage differentiation. Cell. Mol. Life Sci. 68, 3121–3131 (2011). https://doi.org/10.1007/s00018-010-0618-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-010-0618-x