Abstract
Oriented cellulose deposition is critical to plant patterning and models suggest microtubules constrain cellulose synthase movements through the plasma membrane. Though widespread in plants, urochordates are the only animals that synthesize cellulose. We characterized the distinctive cellulose microfibril scaffold of the larvacean house and its interaction with house structural proteins (oikosins). Targeted disruption of cytoskeletal elements, secretory pathways, and plasma membrane organization, suggested a working model for templating extracellular cellulose microfibrils from animal cells that shows both convergence and differences to plant models. Specialized cortical F-actin arrays template microfibril orientation and glycosylphosphatidylinositol-anchored proteins in lipid rafts may act as scaffolding proteins in microfibril elongation. Microtubules deliver and maintain cellulose synthase complexes to specific cell membrane sites rather than orienting their movement through the membrane. Oikosins are incorporated into house compartments directly above their corresponding cellular field of expression and interact with the cellulose scaffold to a variable extent.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cellulose is the most abundant natural product in the biosphere. In plants, cellulose plays a key role in structural support and the oriented deposition of cellulose microfibrils is critical to anisotropic growth during development [1]. Ever since their discovery in plants [2], microtubules have been proposed to control the orientation of cellulose microfibril deposition in the cell wall. Most models suggest microtubules constrain movement of cellulose synthase (CesA) complexes in the membrane [1]. An alternative hypothesis postulates that microtubules regulate microfibril length [3] with longer microfibrils providing greater resistance to cell expansion parallel to fiber orientation, thus generating an anisotropic shaping force. Nevertheless, more than 40 years after formulation of a link between microtubules and cellulose fibril deposition, the molecular nature of this link remains unclear and is debated [1].
The ability to synthesize cellulose is also found in prokaryotes and fungi, but is restricted to the urochordate subphylum in animals. Urochordate CesAs have been identified in two ascidians, Ciona savignyi [4], and C. intestinalis [5, 6]. Ascidians possess only one CesA gene, whereas we recently identified duplicated CesA genes (Od-CesA1 and Od-CesA2) in a larvacean Oikopleura dioica, which have evolved distinct temporal and functional specializations [7]. Phylogenetic analyses indicate that urochordates acquired the CesA gene by horizontal transfer from a prokaryote [4–7]. Since molecular phylogenetic data [8] and a filter-feeding hypothesis on urochordate evolution [9] suggest larvaceans branch nearer the base of the chordate lineage than ascidians and thaliaceans, the findings indicate that the urochordate lineage acquired the CesA gene by a single transfer event prior to divergence into three sister classes. Given this probable origin of urochordate CesAs, we were interested to compare mechanisms of cellulose microfibril orientation in animal cells to those currently elucidated in phylogenetically distant plant cells.
Among the three urochordate classes, ascidians and thaliaceans live inside a unique integument, the tunic, containing crystalline cellulose as a structural component, whereas the larvaceans secrete a complex filter-feeding house that surrounds the animal (Fig. 1a, b). The house is initially secreted as a compact rudiment by a specialized oikoplastic epithelium, and several rudiment layers are often observed stacked above the trunk (Fig. 1c). Upon escape of the animal from an inflated house, the outermost rudiment swells and is subsequently expanded by specific movements of the trunk and tail until the entire animal is contained within the mature structure. In the larvacean house, sinusoidal movements of the tail control water flow. Seawater enters the house through an inlet filter (if) that eliminates large particles and those of appropriate size are then trapped in the food-concentrating filter (fcf) and brought to the mouth, where they are ingested with the aid of a mesh secreted by the endostyle. Filtered seawater flows out the exit spout (Fig. 1b). Of the duplicated CesA genes in O. dioica, spatial-temporal expression analysis indicated that the Od-CesA1 produces long cellulose fibrils along the larval tail, which play a role in notochord and tail morphogenesis, whereas Od-CesA2 is involved in the cellulose scaffold of the post-metamorphic filter-feeding house [7]. Repetitive synthesis (every 4 h) of an identical house structure from a transparent, accessible epithelium makes the process of house construction an excellent animal counterpoint to investigation of this developmentally important phenomenon in plants.
The filter-feeding house of Oikopleura dioica. a O. dioica in a fully expanded filter-feeding house. b Schematic representation of a (modified after [10]). The animal is shown in grey with mouth oriented to the right, the gonad to the left, and the tail projecting downwards. Water flow through the house is indicated with blue arrows and the inlet filter (if) and food-concentrating filter (fcf) are indicated. Food particles collected by the fcf are brought (black arrow) toward the mouth where they are captured by a mesh secreted by the endostyle and then ingested. c A day-5 animal with gonad at top and mouth at bottom shows two non-inflated pre-house rudiments (arrowheads) secreted around the trunk. d The oikoplastic epithelium of a fixed animal was cut ventrally, removed from the trunk, and spread on a slide with the nuclei stained by Hoechst 33258. The anterior-posterior axis is from top to bottom. The fields of Fol and Eisen are indicated by arrows. On the right-hand side of the panel, an image of the right half of the bilaterally symmetric pre-house rudiment has been placed on top of the right side of the bilaterally symmetric epithelial spread. L and U, lower and upper region of the right half of the fcf anlagen that lies directly above the Fol region of the epithelium. The right-side if anlagen of the house rudiment is situated above the Eisen field
Materials and methods
Animal culture
Oikopleura dioica were maintained continuously in culture at 15°C [10].
Confocal analysis of cellulose microfibrils and F-actin filaments
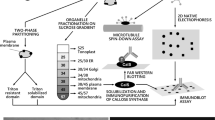
Day 4–5 animals were fixed in 4% paraformaldehyde, 0.1% saponin, 0.1 M MOPS pH 7.5, and 0.5 M NaCl at 4°C overnight. Fixed animals were rinsed with PBS/0.1% saponin/0.1% Tween 20 (S/PBS-T), and then blocked with 3% BSA + S/PBS-T at 4°C overnight. Cellulose content of the house rudiment was probed by incubation in 1% BSA + S/PBS-T containing rCBD-Protein L (10 μg/ml; Fluka) and mouse IgG (10 μg/ml, Sigma) at 4°C overnight, followed by incubation in Rhodamine Red X conjugated goat anti-mouse IgG (1:200 in 1% BSA + S/PBS-T) at 4°C overnight [7]. To analyze cortical F-actin, Alexa Fluor 488 phalloidin (10 units/ml; Molecular Probes) was added to each of the incubation steps. Nuclei were counterstained with 1 µM To-Pro-3 iodide (Molecular Probes). Specimens were mounted in Vectashield (Vector Laboratories) and analyzed with a Leica TCS laser scanning confocal microscope.
Antibodies against oikosins
In addition to the original seven oikosins that were characterized [11, 12], we have now identified an additional 45 oikosins [unpublished observation]. Of the 52 oikosins, six were selected as targets for the generation of polyclonal antibodies. The peptide epitopes used to generate the respective antibodies are given in Table 1. Custom rabbit antisera against oikosins 1 (AJ308491), 2 (AJ308492), and 3 (AJ308495) were made by Washington Biotechnology Inc. (Baltimore, MD) whereas anti-oikosin 8 (FN806849), 18 (FN806850), and 32 (FN806851) rabbit antibodies were generated by 21st Century Biochemicals (Marlborough, MA). In the later case, sera were obtained after five consecutive bleeds and antibodies were affinity-purified from the sera.
Western blotting
To collect house rudiments free of contaminating particles, adult animals (day 4–5) were forced out of their houses and transferred to filtered seawater. House rudiments were removed as soon as the animals started expanding them. Aliquots of 50 house rudiments were centrifuged at 12,000 × g for 10 min at 4°C to remove excess seawater and then stored at –80°C. Extraction of the house proteins was performed as previously [11]. The proteins in the extraction were precipitated with 20% trichloroacetic acid/acetone, rinsed with acetone and diethyl ether, and then re-dissolved in loading buffer. The extract was applied onto SDS–PAGE using 5–20% (oikosin 1), 12.5% (oikosins 2 and 3) or 15% (oikosins 8, 18 and 32) acrylamide gels, and transferred onto PVDF membranes (Millipore). Membranes were blocked in 5% skim milk/PBS-T for 1 h at room temperature. Target proteins were probed with anti-oikosin 1, 2, and 3 rabbit sera (1:1,000) or anti-oikosin 8, 18, 32 rabbit antibodies (1:1,000) in the blocking solution overnight at 4°C. Membranes were washed three times in PBS-T, and then incubated with anti-rabbit IgG (Jackson Immunoresearch) conjugated HRP (1:15,000) for 1 h at room temperature. Membranes were washed three times in PBS-T, and processed using ECL + (GE Healthcare).
In situ hybridizations and immunostainings
Whole mount in situ hybridizations for oikosins and fluorescent whole mount in situ hybridization for CesA2 were performed as previously [7, 13] using probes described in Supplementary Table 1. To localize oikosins in the pre-house structure, day 4 animals were fixed in 4% paraformaldehyde/0.1 M MOPS pH 7.5/0.5 M NaCl at 4°C overnight. Fixed animals were rinsed with PBS/0.1% Tween 20 (PBS-T), and then blocked with 3% BSA + PBS-T at 4°C overnight. Samples were incubated at 4°C overnight with antisera (1:100 dilution) for oikosin 1–3, and affinity purified antibodies (1:100 dilution) for oikosin 8, 18, and 32 in 3% BSA + PBS-T, washed with PBS-T, post-fixed in 4% paraformaldehyde/0.1 M MOPS pH 7.5/0.5 M NaCl for 30 min at RT, washed as after the first fixation and incubated at 4°C overnight with anti-rabbit IgG conjugated Alexa 568 (1:1,000) in 3% BSA + PBS-T. Samples were washed with PBS-T, incubated 15 min in 1 μM To-Pro-3 iodide to counterstain DNA, and then washed with PBS-T. To analyze cortical F-actin, Alexa Fluor 488 phalloidin (10 units/ml; Molecular Probes) was added to each of incubation steps. Specimens were mounted in Vectashield and analyzed with a Leica TCS laser scanning confocal microscope.
Incubation of animals with anti-oikosin antibodies
Lots of ten day-4 animals were placed into 10-ml volumes of seawater containing respective anti-oikosin (1, 2, 3, 8, 18, and 32) antibodies at 1:100 dilutions, and incubated for 12 h at room temperature with gentle agitation.
Inhibitor assays
Day-3–5 animals (30–50 animals) were transferred into 800 ml of fresh seawater without food containing one of the inhibitory reagents (all from Sigma, except lumicolchicine from ChromaDex Inc.). Dose-response assays to determine effective inhibitor concentrations are summarized in Table 2. Final working concentrations for more detailed analyses were: cytochalasin B (0.8 μM), colchicine (25 μM), lumicolchicine (25 μM), monensin (32 nM), filipin III (0.15 μM), or neomycin (100 μM). Animals were cultured at these concentrations for 4–12 h at room temperature. Final inhibitor concentrations were prepared from stock solutions of 20 mM cytochalasin B in DMSO, 100 mM colchicine in seawater, 50 mM lumicolchicine in ethanol, 7.6 mM filipin III in DMSO, 10 mM monensin in ethanol, and 10 mM neomycin in seawater. To determine whether DMSO or ethanol solvents had any effect on house structure, experiments in which animals were incubated in the corresponding concentrations of the solvents alone were also carried out.
Results and discussion
Components of the filter-feeding house
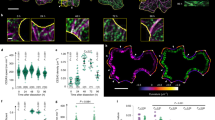
In O. dioica, the fcf and if are readily identified in pre-house rudiments and superimpose onto the oikoplastic epithelial fields of Fol and Eisen, respectively (Fig. 1d), which secrete components that are specifically incorporated into these filters [11, 12]. The extracellular cellulose microfibrils produced by cellulose synthase complexes encoded by the Od-CesA2 gene are involved in forming a scaffold of the filter-feeding house [7]. Using rCBD-Protein L, we revealed a cellulose-based skeletal structure of the house rudiment of a representative larvacean, O. dioica (Fig. 2a). The fcf was composed of two layers with different mesh sizes, designated U and L (Fig. 2b). Cellulose microfibrils in the L-region were secreted from the anterior Fol to form a fine mesh. The U-region mesh was characterized by a transverse array of filaments secreted by two sets of seven cells flanking the posterior Fol region. The if, secreted above the Eisen region, exhibited a meshwork composed of a single warp and double weft thread (Fig. 2c). The termini of each cellulose bundle branched into smaller fibrils that projected to smaller cells surrounding the giant Eisen cells. The weft thread microfibrils of the if superpose above a transverse apical array of F-actin filaments in the three central giant Eisen cells.
Cellulose-based scaffold of the filter-feeding house. a Confocal image stack of cellulose microfibrils in the pre-house rudiment (upper) superimposed on To-Pro-3 stained nuclei (blue, lower) of the oikoplastic epithelium responsible for secretion of house components. b Confocal image stacks of cellulose microfibrils in the fcf of the rudiment (upper left) showing nascent fibrils for the L and U regions. Fibrils were secreted from distinct cells in the Fol region of the epithelium (red outlines) on a lateral view of the Hoechst-stained animal. Scale bars indicate 20 μm. c Confocal image stacks of the orientation of cellulose microfibrils in the maturing if (upper left). The three remaining images show the nascent if where cellulose weft microfibrils are oriented above a transverse apical network of cellular F-actin in the three central giant Eisen cells. Cortical F-actin and stained nuclei indicate the positions of individual cells in the Eisen region. Scale bars indicate 20 μm. The images in the right panels show three-fold zooms of regions indicated by dashed squares (1, 2, 3) in the left and central panels. The pairs of cellulose fibrils in the doublet-weft threads appear to flank the transverse actin fibers
Previous analyses indicated that the house consists of at least 20–30 polypeptides, a number of which are heavily glycosylated [11]. Through protein microsequencing [11, unpublished observation] and representation difference analysis [12], at least 50 genes (oikosins) with distinct, complementary expression patterns restricted to defined regions of the oikoplastic epithelium have been identified. We have now developed antibodies against six of the protein products: anti-oikosins 1, 2, 3, 8, 18, and 32. The antibodies recognize the targeted oikosins on both Western blots (Fig. 3) and whole mount immunofluorescent staining (Fig. 4). Each of the oikosin proteins was incorporated into the pre-house rudiment directly above the cellular fields from which they were produced and no significant migration in the extracellular space was observed for any of the six oikosins tested. Double labeling of cellulose microfibrils and oikosins revealed that oikosins co-localized with cellulose fibrils to a variable extent, with oikosin 2 showing the most complete co-localization, appearing to coat the cellulose microfibers produced in the U compartment of the fcf (Fig. 5). Thus, cellulose microfibers of varying thickness and orientation are produced in the house by a single CesA2 gene. Different oikosins, produced by specific fields of epithelial cells, interact locally with the variable cellulose scaffold to form the defined compartments of the house. Oikosins 1, 2, 3, 8, and 18 are incorporated into the fcf, whereas the distribution of oikosin 32 is consistent with a role in the house shell rather than in filter meshes.
Western blotting of anti-oikosins. a Pre-house rudiment extracts were run on SDS-PAGE using 5–20% (oikosin 1), 12.5% (oikosins 2 and 3) or 15% (oikosins 8, 18, and 32) acrylamide gels. Predicted molecular weights (kDa) were, oikosin 1, 256; oikosin 2, 84; oikosin 3, 67; oikosin 8, 34, oikosin 18, 26, and oikosin 32, 47. With the exception of anti-oikosin 1 and 8, applied antibodies detected single bands corresponding to expected molecular weights. The band (25 kDa) detected by anti-oikosin-8 antibody is smaller than calculated size (34 kDa), but a mass spectrometric sequenced peptide from the same-sized 25-kDa band is a perfect match to part of the oikosin 8 amino acid sequence. Therefore, oikosin 8 may be post-translationally modified or migrate anonymously. b Anti-oikosin-1 detected several protein bands in the 40 to 50-kDa range in addition to the predicted band at 260 kDa, possibly indicative of some degradation or post-translational processing of the full length oikosin 1 protein detected at the top of the blot. Confocal image stacks of oikosin 1 immunostaining revealed signal as expected in the seven giant Fol cells and in the pre-house rudiment directly above these giant cells as would be expected based on the in situ pattern for oikosin 1 gene expression (see Fig. 4). However, signals were also detected in the four peripheral giant cells in the Eisen field, where no oikosin 1 mRNA in situ signal is ever detected. This suggests that the antibody also recognizes a similar domain in an additional protein(s) and this may also account for some of the staining in the 40 to 50-kDa range. Scale bar indicates 100 µm
Immunostaining patterns for oikosins 1, 2, 3, 8, 18, and 32. In situ expression patterns for the genes coding the six oikosins are summarized as digital expression (exp) domains on the Hoechst-stained epithelial spreads. Spreads are oriented as in Fig. 1d. Actual in situ patterns are shown for oikosins 1, 2, and 3 in [11] and for oikosins 8, 18, and 32 in supplementary Fig. 1. All immunostaining images show lateral views of the trunk, with the mouth towards the upper left. The entire Fol and Eisen regions are indicated on the paired images for Oikosin 32. Sub-regions of the Fol are designated on other images: af, anterior Fol; g, giant Fol; n, nasse cells; pf, posterior Fol. Oikosin 1 and 18 genes are expressed in the 14 giant Fol cells. Oikosin 3 and 8 genes are expressed in the anterior Fol. Oikosin 2 is expressed in several rows of perioral cells in the anterior part of the epithelium, a single row of cells surrounding the anterior Fol, the three rows of small nasse cells, the posterior Fol and the three central giant Eisen cells. Oikosin 32 is expressed in the posterior dorsal part of the epithelium and in bands extending ventrally. In the immunoflourescent images, oikosin proteins are stained in green, actin in red, and DNA in blue. Scale bars indicate 100 µm
Co-staining patterns for oikosins and cellulose fibrils. Confocal image stacks of oikosins (red) and cellulose (green) in the house rudiment are shown. L and U, lower and upper region of the fcf anlagen. Scale bars indicate 100 μm. Zooms (1.6-, 4.8-, 2.3-, 6.8-, 3.5-. and 10-fold, from top to bottom) show the extent to which respective oikosins co-localize with cellulose microfibirils
We further attempted to determine if house formation could be disrupted by incubations of the animals in the presence of anti-oikosin antibodies. Under these conditions, no alteration of house structures was observed and animals developed and grew normally. It may be that polyclonal antibodies raised against entire oikosin proteins would be more effective in this strategy than the currently available antibodies raised against defined, short, peptides within each oikosin.
Cytoskeleton-mediated templating of house structures
In distinct contrast to prevailing models in plant cells, confocal image stacks of the nascent if and the underlying giant Eisen cells showed a clear parallelism between nascent cellulose weft fibers and an underlying transverse array of cortical F-actin filaments (Fig. 2c). To assess roles that cytoskeletal and cell membrane components might play in templating the O. dioica house, we employed an array of inhibitors. Dose-response assays were carried out for each inhibitor to determine the effective concentration range (Table 2).
When day 5 animals were cultured for several hours in the presence of 0.8 μM cytochalasin B, a specific disrupter of F-actin formation, the structure of the fcf was clearly perturbed (Fig. 6a). The sheath surrounding the fcf appeared normal but the ordered interior ribbed meshwork was largely absent. At 15°C, O. dioica produces one new house every 4 h throughout the post-metamorphic portion of the life cycle. Thus, the effect of F-actin disruption on house structure occurred within one house synthetic cycle. The effect of this disruption on fcf morphology was reversible (Fig. 6b). When animals were cultured in cytochalasin B for 6 h to disrupt fcf structure and then transferred to seawater in the absence of inhibitor, normal filter morphology was recovered in subsequently synthesized houses. Confocal image stacks of the nascent if and underlying Eisen cells revealed that 0.8 μM of cytochalasin B was sufficient to specifically disrupt the apical transverse array of F-actin that paralleled weft thread microfibril deposition while leaving the basic cellular cortical actin network underlying the cell membrane intact (Fig. 7). Specific disruption of the transverse F-actin array resulted in concomitant disruption of the meshwork of overlying cellulose microfibrils, implicating this array in their ordered deposition. A parallel orientation of extracellular cellulose microfibrils and cortical F-actin has also been noted in the brown algae Dictyota dichotoma [14].
Effects of disrupting cellular actin filaments on the structure of the fcf. a Day 5 animals cultured in seawater in the presence of 0.8 μM cytochalasin B (CB) for 6 h to disrupt the cellular F-actin network show obvious morphological defects in the fcf compared to control animals cultured in the presence of the inhibitor solvent, 0.004% DMSO, alone (DMSO). The characteristic ribbed structure of the fcf was no longer present. Scale bars indicate 500 µm. b The effect of cytochalasin B on house structure was reversible by subsequent 6 h culture of animals in the absence of this inhibitor
Typical phenotypes of nascent inlet filters affected by drugs targeting various cellular components. Day-5 animals were cultured in seawater in the presence of each of the drugs and the effects were assayed by confocal microscopy on the F-actin network (left column) and the nascent cellulose microfibril distribution (central column). These two staining patterns were superposed with DNA counterstaining (right column). Insets are close-up images (1.9-, 2.5-, 2.9-, 2.8-fold from top to bottom) of areas indicated by dotted squares in each panel. Concentrations of inhibitor and incubation times were: cytochalasin B, 0.8 μM, 6 h; colchicine, 25 μM, 4 h; filipin III, 0.15 μM, 8 h; monensin, 32 nM, 6 h. The control configurations for F-actin and cellulose microfibril deposition are shown in Fig. 2c. The scale bar on the upper left applies to all images
We next examined the effects of microtubule disruption on if morphology. Culture of animals in 25 μM of colchicine had no visible effect on the transverse array of cortical F-actin but did induce a non-uniform width in the size of thread bundles as well as non-uniform spacing between adjacent thread lines (Fig. 7). These effects were observed in both weft- and warp-thread lines. At similar concentrations (25–50 μM), the inactive analog lumicolchicine did not show the same disruption of microfibril patterning, though at 100 μM, epithelial cells detached and became necrotic, suggesting a non-specific general toxic effect of this analog at higher concentrations (Supplementary Fig. 2).
Plant cellulose microfibrils are produced by a terminal multimeric complex of CesA subunits on the plasma membrane [15], and in ascidians, terminal complexes on the plasma membrane of epidermal cells face the tunic [16, 17]. In developing xylem of Arabidopsis, cortical microtubules are continually required to maintain correct CesA localization [18]. Unfortunately, little is known concerning CesA complex turnover in the membrane, with only one estimate of 20 min available for the moss protonemata [19]. Our results are consistent with a patchy absence of synthase complexes in the membrane, because of a failure of the disrupted microtubule network to deliver/maintain these complexes at correct locations in the plasma membrane.
Recently, the glycosylphosphatidylinositol (GPI)-anchored protein, COBRA, has been shown to play an important role in cellulose microfibril orientation in Arabidopsis [20] and mutations at five loci in the PEANUT1-5 locus of Arabidopsis, which cause defects in GPI-anchor synthesis, result in aberrant cell wall synthesis with decreased incorporation of crystalline cellulose [21]. GPI-anchored proteins are found in lipid rafts [22] and filipin III, a fungal polyene macrolide, is known to disrupt these rafts. The phenotype observed when animals were cultured in 0.15 µM of filipin III was a reduced length of warp thread lines (Fig. 7) perpendicular to the transverse cortical F-actin array. No disruption of the spacing between microfibrils was detected. The most common point of microfibril arrest was at the juncture between the first and the second of the three central giant Eisen cells. GPI-anchored proteins are known to move between adjacent cell membranes in a process mediated by lipid rafts [23] and our results suggest that this is integral to elongation of warp thread cellulose microfibrils in construction of the nascent if. It is unclear if this occurs via direct or indirect scaffolding of cellulose microfibrils by GPI-anchored proteins. GPI-anchored and other putative scaffolding proteins involved in ordered cellulose microfibril deposition are for the most part likely to transit the Golgi. Culture of animals in 32 nM of monensin, an inhibitor of intra-Golgi trafficking, disrupted cellulose microfibril elongation in both weft and warp threads, though again, there was no effect on inter-fiber spacing in either direction. Under these conditions, it is possible that CesA complexes remained correctly positioned and spaced in the membrane but that linkage of the nascent cellulose microfibrils to scaffolding proteins for elongation is critical to a feedback loop of continued cellulose synthesis and that putative scaffolding proteins had higher turnover rates than the synthase complexes in the membrane.
Pre-house release and inflation
Involvement of GPI-anchored proteins in directly or indirectly linking the nascent pre-house rudiment to the plasma membrane of the underlying oikoplastic epithelium also suggests regulated activity of phospholipases known to cleave GPI anchors as a possible mechanism for the cyclic release of this templated structure from the underlying cellular monolayer. To test for a potential role of phospholipase action on GPI-anchored proteins during pre-house release and inflation, the effect of neomycin, an inhibitor of both phospholipase C and D, was assessed on day 3 animals. Normally, the animals leave their expanded houses approximately every 4 h and readily inflate a new house from an earlier secreted underlying pre-house rudiment. Neomycin disrupted the expansion of pre-houses and led to the accumulation of non-inflated house rudiments over time (Fig. 8). The accumulated pre-house rudiments appeared structurally normal. The effect of neomycin was immediate as animals that had just abandoned their houses and were then transferred to seawater containing neomycin were unable to expand existing pre-house rudiments. After approximately 3.5 h in the presence of 100 μM of neomycin, all animals had lost their ability to expand houses. At this time, 90% of the animals (n = 500) remained viable. When the animals were then transferred to fresh seawater, 87% of them (n = 450) were able to inflate houses within 3.5 h of transfer, demonstrating clear reversibility of the effect of neomycin. These results are consistent with possible roles for phospholipases in cleaving GPI anchors to promote pre-house release and inflation.
Neomycin inhibits house release and inflation. a, c Two secreted pre-houses (arrowheads) are located on the trunks of control animals in seawater. b, d In the presence of 100 μM neomycin for 4 h, four pre-house rudiments (arrowheads) accumulated on the trunks and house inflation was inhibited. The distal rudiment layers were less tightly associated with the epithelium and tended to be dislocated upon manipulation, though remaining attached at what appear to be hinge points located anteroventrally on the trunk. a, b Transmission images. c, d Confocal image stacks: cellulose (green); actin (red), and DNA (blue). Scale bars indicate 20 μm
Conclusions
In Fig. 9, we present a working model of cytoskeletal-mediated cellulose microfibril deposition consistent with our experimental results. While there are points of convergence with favored models of cellulose deposition in plant cells, there are also key differences. This probably reflects that pathways for templating extracellular cellulose-based structures in plants and animals are likely to be analogous rather than homologous. We propose that in the O. dioica if, F-actin filaments, rather than microtubules, template cellulose microfibril orientation and that fibril elongation does not necessarily implicate movement of CesA complexes through the membrane but can occur through attachment of nascent fibrils to scaffolding proteins anchored in lipid rafts. This includes elongation of fibrils across membranes of adjacent cells.
Hypothetical model for the ordered incorporation of cellulose microfibrils into the inlet filter of the Oikopleura dioica house. Directions of weft and warp threads of the mesh, corresponding to that defined in Fig. 2c, are indicated by green double-headed arrows. The apical region of one of three central giant Eisen cells is shown abutting a second giant cell and surrounding cells of the Eisen field. The fluorescent in situ image at the bottom left shows that the cellulose synthase gene (CesA2) is not expressed in the giant cells themselves but rather in smaller cells surrounding the giant Eisen field (green, CesA2 mRNA; red, DNA; dashed outline indicates the giant Eisen field; scale bar indicates 20 µm). The apical cortical array of F-actin in giant cells guides the orientation of weft-thread elongation, either via transmembrane scaffolding proteins or complexes that link extracellular scaffolding proteins to membrane proteins organized by the underlying F-actin network. Microtubules in surrounding cells sort the CesA units to correct sites on plasma membrane where microfibril synthesis originates. Lipid rafts in the apical giant cell plasma membrane surface are involved in warp-thread elongation across adjacent cells. Golgi-sorted proteins provide the links between transverse F-actin fibres, lipid rafts, and cellulose microfibril orientation
Anti-oikosin antibody staining patterns in pre-house rudiments suggest that as a general rule, these structural proteins are incorporated into house compartments directly above their corresponding cellular field of expression, and show little significant migration in the extracellular space. They interact with the cellulose microfibril scaffold to a variable extent, with some, such as oikosin 2, essentially coating the cellulose microfibrils. Transient expression of oikosin-fluorescent protein fusion plasmid constructs driven by endogenous promoters is now feasible in O. dioica [24] and could assist in assessing the distribution and functional roles of a wider panel of oikosins in the house. Promoter swapping and capped mRNA experiments could be used to examine the effects of spatial mis-expression of oikosins on house structure. However, mosaicism could complicate interpretation of most such experiments and it would be preferable to execute these studies once stable transformant strategies have been established in this emerging model organism.
The impacts of lipid raft disruption on cellulose scaffold elaboration and the inhibition of house rudiment inflation by neomycin implicate GPI-anchored proteins in templating the house rudiment and regulating its release from the epithelium and expansion into a fully functional feeding structure. It would be of interest to perform proteomic screens of oikoplastic epithelial membrane preparations or in situ screening of in silico identified GPI-anchored proteins in the O. dioica genome to isolate candidate GPI-anchored proteins involved in these processes and to determine to what extent, if any, homologies exist to GPI-anchored proteins involved in scaffolding plant cellulose-based extracellular matrices. With sequencing of its compact 70 Mb [25] genome nearing completion, O. dioica provides an excellent comparative animal model to active research in plants aimed at further understanding cellular mechanisms for weaving the abundant natural product, cellulose, into an array of elegant and functional extracellular structures.
References
Smith LG, Oppenheimer DG (2005) Spatial control of cell expansion by the plant cytoskeleton. Annu Rev Cell Dev Biol 21:271–295
Ledbetter MC, Porter KR (1963) A “microtubule” in plant cell fine structure. J Cell Biol 19:239–250
Wasteneys GO (2004) Progress in understanding the role of microtubules in plant cells. Curr Opin Plant Biol 7:651–660
Matthysse AG, Deschet K, Williams M, Marry M, White AR, Smith WC (2004) A functional cellulose synthase from ascidian epidermis. Proc Natl Acad Sci USA 101:986–991
Nakashima K, Yamada L, Satou Y, Azuma J, Satoh N (2004) The evolutionary origin of animal cellulose synthase. Dev Genes Evol 214:81–88
Sasakura Y, Nakashima K, Awazu S, Matsuoka T, Nakayama A, Azuma J-I, Satoh N (2005) Transposon-mediated insertional mutagenesis revealed the functions of animal cellulose synthase in the ascidian Ciona intestinalis. Proc Natl Acad Sci USA 102:15134–15139
Sagane Y, Zech K, Bouquet J-M, Schmid M, Bal U, Thompson EM (2010) Functional specialization of cellulose synthase genes of prokaryotic origin in chordate larvaceans. Development 137:1483–1492
Delsuc F, Tsagkogeorga G, Lartillot N, Philippe H (2008) Additional molecular support for the new chordate phylogeny. Genesis 46:592–604
Satoh N (2009) An advanced filter-feeder hypothesis for urochordate evolution. Zool Sci 26:97–111
Bouquet J-M, Spriet E, Troedsson C, Otterå H, Chourrout D, Thompson EM (2009) Culture optimization for the emergent zooplanktonic model organism Oikopleura dioica. J Plankton Res 31:359–370
Spada F, Steen H, Troedsson C, Kallesøe T, Spriet E, Mann M, Thompson EM (2001) Molecular patterning of the oikoplastic epithelium of the larvacean tunicate Oikopleura dioica. J Biol Chem 276:20624–20632
Thompson EM, Kallesøe T, Spada F (2001) Diverse genes expressed in distinct regions of the trunk epithelium define a monolayer cellular template for construction of the oikopleurid house. Dev Biol 238:260–273
Seo HC, Edvardsen RB, Maeland AD, Bjordal M, Jensen MF, Hansen A, Flaat M, Reinhardt R, Chourrout D (2004) Hox cluster disintegration with persistent anteroposterior order of expression in Oikopleura dioica. Nature 431:67–71
Katsaros C, Karyophyllis D, Galatis B (2003) F-actin cytoskeleton and cell wall morphogenesis in brown algae. Cell Biol Int 27:209–210
Kimura S, Laosinchai W, Itoh T, Cui X, Linder CR, Brown RM Jr (1999) Immunogold labeling of rosette terminal cellulose-synthesizing complexes in the vascular plant Vigna angularis. Plant Cell 11:2075–2085
Kimura S, Itoh T (1996) New cellulose synthesizing complexes (terminal complexes) involved in animal cellulose biosynthesis in the tunicate Metandrocarpa uedai. Protoplasma 194:151–163
Kimura S, Itoh T (2004) Cellulose synthesizing terminal complexes in the ascidian. Cellulose 11:377–383
Gardiner JC, Taylor NG, Turner SR (2003) Control of cellulose synthase complex localization in developing xylem. Plant Cell 15:1740–1748
Rudolph U, Schnepf E (1988) Investigations of the turnover of the putative cellulose-synthesizing particle ‘rosettes’ within the plasma membrane of Funaria hygrometrica protonema cells. I. Effects of monensin and cytochalasin B. Protoplasma 143:63–73
Roudier F, Fernandez AG, Fujita M, Himmelspach R, Borner GH, Schindelman G, Song S, Baskin TI, Dupree P, Wasteneys GO, Benfey PN (2005) COBRA, an Arabidopsis extracellular glycosylphopatidylinositol-anchored protein, specifically controls highly anisotropic expansion through its involvement in cellulose microfibril orientation. Plant Cell 17:1749–1763
Gillmor CS, Lukowitz W, Brininstool G, Sedbrook JC, Hamann T, Poindexter P, Somerville C (2005) Glycosylphosphatidylinositol-anchored proteins are required for cell wall synthesis and morphogenesis in Arabidopsis. Plant Cell 17:1128–1140
Mayor S, Rao M (2004) Rafts: scale-dependent, active lipid organization at the cell surface. Traffic 5:231–240
Liu T, Li R, Pan T, Liu D, Petersen RB, Wong BS, Gambetti P, Sy MS (2002) Intercellular transfer of the cellular prion protein. J Biol Chem 277:47671–47678
Clarke T, Bouquet JM, Fu X, Kallesøe T, Thompson EM (2007) Rapidly evolving lamins in a chordate, Oikopleura dioica, with unusual nuclear architecture. Gene 396:159–169
Seo HC, Kube M, Edvardsen RB, Jensen MF, Beck A, Spriet E, Gorsky G, Thompson EM, Lehrach H, Reinhardt R, Chourrout D (2001) Miniature genome in the marine chordate Oikopleura dioica. Science 294:2506
Acknowledgments
We thank the staff from Appendic Park for supplying animals. This work was supported by grant 17541/S10 NFR-FUGE from the Norwegian Research Council (E.M.T.).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Sagane, Y., Hosp, J., Zech, K. et al. Cytoskeleton-mediated templating of complex cellulose-scaffolded extracellular structure and its association with oikosins in the urochordate Oikopleura . Cell. Mol. Life Sci. 68, 1611–1622 (2011). https://doi.org/10.1007/s00018-010-0547-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-010-0547-8