Abstract
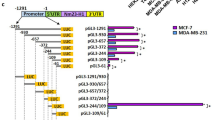
We identified CREB3 as a novel HDAC3-interacting protein in a yeast two-hybrid screen for HDAC3-interacting proteins. Among all class I HDACs, CREB3 specifically interacts with HDAC3, in vitro and in vivo. HDAC3 efficiently inhibited CREB3-enhanced NF-κB activation, whereas the other class I HDACs did not alter NF-κB-dependent promoter activities or the expression of NF-κB target genes. Importantly, both knock-down of CREB3 and overexpression of HDAC3 suppressed the transcriptional activation of the novel CREB3-regulated gene, CXCR4. Furthermore, CREB3 was shown to bind to the CRE element in the CXCR4 promoter and to activate the transcription of the CXCR4 gene by causing dissociation of HDAC3 and subsequently increasing histone acetylation. Importantly, both the depletion of HDAC3 and the overexpression of CREB3 substantially increased the migration of MDA-MB-231 metastatic breast cancer cells. Taken together, these findings suggest that HDAC3 selectively represses CREB3-mediated transcriptional activation and chemotactic signalling in human metastatic breast cancer cells.





Similar content being viewed by others
References
Emiliani S, Fischle W, Van Lint C, Al-Abed Y, Verdin E (1998) Characterization of a human RPD3 ortholog, HDAC3. Proc Natl Acad Sci U S A 95(6):2795–2800
Evans PM, Zhang W, Chen X, Yang J, Bhakat KK, Liu C (2007) Kruppel-like factor 4 is acetylated by p300 and regulates gene transcription via modulation of histone acetylation. J Biol Chem 282(47):33994–34002
Zhang W, Kadam S, Emerson BM, Bieker JJ (2001) Site-specific acetylation by p300 or CREB binding protein regulates erythroid Kruppel-like factor transcriptional activity via its interaction with the SWI-SNF complex. Mol Cell Biol 21(7):2413–2422
Zhang X, Ozawa Y, Lee H, Wen YD, Tan TH, Wadzinski BE, Seto E (2005) Histone deacetylase 3 (HDAC3) activity is regulated by interaction with protein serine/threonine phosphatase 4. Genes Dev 19(7):827–839
Qi D, Bergman M, Aihara H, Nibu Y, Mannervik M (2008) Drosophila Ebi mediates snail-dependent transcriptional repression through HDAC3-induced histone deacetylation. EMBO J 27(6):898–909
Karagianni P, Wong J (2007) HDAC3: taking the SMRT-N-CoRrect road to repression. Oncogene 26(37):5439–5449
Yoon HG, Chan DW, Huang ZQ, Li J, Fondell JD, Qin J, Wong J (2003) Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. EMBO J 22(6):1336–1346
Yoon HG, Wong J (2006) The corepressors silencing mediator of retinoid and thyroid hormone receptor and nuclear receptor corepressor are involved in agonist- and antagonist-regulated transcription by androgen receptor. Mol Endocrinol 20(5):1048–1060
Hodgson MC, Astapova I, Cheng S, Lee LJ, Verhoeven MC, Choi E, Balk SP, Hollenberg AN (2005) The androgen receptor recruits nuclear receptor CoRepressor (N-CoR) in the presence of mifepristone via its N and C termini revealing a novel molecular mechanism for androgen receptor antagonists. J Biol Chem 280(8):6511–6519
Sankar N, Baluchamy S, Kadeppagari RK, Singhal G, Weitzman S, Thimmapaya B (2008) p300 provides a corepressor function by cooperating with YY1 and HDAC3 to repress c-Myc. Oncogene 27(43):5717–5728
Choo MK, Yeo H, Zayzafoon M (2009) NFATc1 mediates HDAC-dependent transcriptional repression of osteocalcin expression during osteoblast differentiation. Bone 45(3):579–589
Feng Y, Wang X, Xu L, Pan H, Zhu S, Liang Q, Huang B, Lu J (2009) The transcription factor ZBP-89 suppresses p16 expression through a histone modification mechanism to affect cell senescence. FEBS J 276(15):4197–4206
Togi S, Kamitani S, Kawakami S, Ikeda O, Muromoto R, Nanbo A, Matsuda T (2009) HDAC3 influences phosphorylation of STAT3 at serine 727 by interacting with PP2A. Biochem Biophys Res Commun 379(2):616–620
Das C, Kundu TK (2005) Transcriptional regulation by the acetylation of nonhistone proteins in humans—a new target for therapeutics. IUBMB Life 57(3):137–149
Viatour P, Legrand-Poels S, van Lint C, Warnier M, Merville MP, Gielen J, Piette J, Bours V, Chariot A (2003) Cytoplasmic IkappaBalpha increases NF-kappaB-independent transcription through binding to histone deacetylase (HDAC) 1 and HDAC3. J Biol Chem 278(47):46541–46548
Gao Z, Chiao P, Zhang X, Lazar MA, Seto E, Young HA, Ye J (2005) Coactivators and corepressors of NF-kappaB in IkappaB alpha gene promoter. J Biol Chem 280(22):21091–21098
Blanco-Garcia N, Asensio-Juan E, de la Cruz X, Martinez-Balbas MA (2009) Autoacetylation regulates P/CAF nuclear localization. J Biol Chem 284(3):1343–1352
Freiman RN, Herr W (1997) Viral mimicry: common mode of association with HCF by VP16 and the cellular protein LZIP. Genes Dev 11(23):3122–3127
Burbelo PD, Kozak CA (1998) Mapping of the murine LZIP gene (Creb3) to chromosome 4. Genomics 54(2):357–358
Luciano RL, Wilson AC (2000) N-terminal transcriptional activation domain of LZIP comprises two LxxLL motifs and the host cell factor-1 binding motif. Proc Natl Acad Sci U S A 97(20):10757–10762
Audas TE, Li Y, Liang G, Lu R (2008) A novel protein, Luman/CREB3 recruitment factor, inhibits Luman activation of the unfolded protein response. Mol Cell Biol 28(12):3952–3966
Luciano RL, Wilson AC (2002) An activation domain in the C-terminal subunit of HCF-1 is important for transactivation by VP16 and LZIP. Proc Natl Acad Sci U S A 99(21):13403–13408
Sung HJ, Kim YS, Kang H, Ko J (2008) Human LZIP induces monocyte CC chemokine receptor 2 expression leading to enhancement of monocyte chemoattractant protein 1/CCL2-induced cell migration. Exp Mol Med 40(3):332–338
Suzuki M, Shinohara F, Sato K, Taniguchi T, Takada H, Rikiishi H (2003) Interleukin-1beta converting enzyme subfamily inhibitors prevent induction of CD86 molecules by butyrate through a CREB-dependent mechanism in HL60 cells. Immunology 108(3):375–383
Canettieri G, Morantte I, Guzman E, Asahara H, Herzig S, Anderson SD, Yates JR 3rd, Montminy M (2003) Attenuation of a phosphorylation-dependent activator by an HDAC-PP1 complex. Nat Struct Biol 10(3):175–181
Yoon HG, Choi Y, Cole PA, Wong J (2005) Reading and function of a histone code involved in targeting corepressor complexes for repression. Mol Cell Biol 25(1):324–335
Jang SW, Kim YS, Lee YH, Ko J (2007) Role of human LZIP in differential activation of the NF-kappaB pathway that is induced by CCR1-dependent chemokines. J Cell Physiol 211(3):630–637
Jang SW, Kim YS, Kim YR, Sung HJ, Ko J (2007) Regulation of human LZIP expression by NF-kappaB and its involvement in monocyte cell migration induced by Lkn-1. J Biol Chem 282(15):11092–11100
Matteucci E, Ridolfi E, Maroni P, Bendinelli P, Desiderio MA (2007) c-Src/histone deacetylase 3 interaction is crucial for hepatocyte growth factor dependent decrease of CXCR4 expression in highly invasive breast tumor cells. Mol Cancer Res 5(8):833–845
Nicolas E, Ait-Si-Ali S, Trouche D (2001) The histone deacetylase HDAC3 targets RbAp48 to the retinoblastoma protein. Nucleic Acids Res 29(15):3131–3136
Liang Z, Wu H, Reddy S, Zhu A, Wang S, Blevins D, Yoon Y, Zhang Y, Shim H (2007) Blockade of invasion and metastasis of breast cancer cells via targeting CXCR4 with an artificial microRNA. Biochem Biophys Res Commun 363(3):542–546
Sun R, Gao P, Chen L, Ma D, Wang J, Oppenheim JJ, Zhang N (2005) Protein kinase C zeta is required for epidermal growth factor-induced chemotaxis of human breast cancer cells. Cancer Res 65(4):1433–1441
Monte M, Simonatto M, Peche LY, Bublik DR, Gobessi S, Pierotti MA, Rodolfo M, Schneider C (2006) MAGE-A tumor antigens target p53 transactivation function through histone deacetylase recruitment and confer resistance to chemotherapeutic agents. Proc Natl Acad Sci U S A 103(30):11160–11165
Choi KC, Jung MG, Lee YH, Yoon JC, Kwon SH, Kang HB, Kim MJ, Cha JH, Kim YJ, Jun WJ, Lee JM, Yoon HG (2009) Epigallocatechin-3-gallate, a histone acetyltransferase inhibitor, inhibits EBV-induced B lymphocyte transformation via suppression of RelA acetylation. Cancer Res 69(2):583–592
Tabata T, Kokura K, Ten Dijke P, Ishii S (2009) Ski co-repressor complexes maintain the basal repressed state of the TGF-beta target gene, SMAD7, via HDAC3 and PRMT5. Genes Cells 14(1):17–28
Yuan LW, Gambee JE (2001) Histone acetylation by p300 is involved in CREB-mediated transcription on chromatin. Biochim Biophys Acta 1541(3):161–169
Ko J, Jang SW, Kim YS, Kim IS, Sung HJ, Kim HH, Park JY, Lee YH, Kim J, Na DS (2004) Human LZIP binds to CCR1 and differentially affects the chemotactic activities of CCR1-dependent chemokines. FASEB J 18(7):890–892
Su YC, Wu MT, Huang CJ, Hou MF, Yang SF, Chai CY (2006) Expression of CXCR4 is associated with axillary lymph node status in patients with early breast cancer. Breast 15(4):533–539
Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, Varambally S, Ghosh D, Chinnaiyan AM (2007) Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 9(2):166–180
Burger JA, Kipps TJ (2006) CXCR4: a key receptor in the crosstalk between tumor cells and their microenvironment. Blood 107(5):1761–1767
Smith MC, Luker KE, Garbow JR, Prior JL, Jackson E, Piwnica-Worms D, Luker GD (2004) CXCR4 regulates growth of both primary and metastatic breast cancer. Cancer Res 64(23):8604–8612
Yasuoka H, Tsujimoto M, Yoshidome K, Nakahara M, Kodama R, Sanke T, Nakamura Y (2008) Cytoplasmic CXCR4 expression in breast cancer: induction by nitric oxide and correlation with lymph node metastasis and poor prognosis. BMC Cancer 8:340
Mariadason JM (2008) Dissecting HDAC3-mediated tumor progression. Cancer Biol Ther 7(10):1581–1583
Acknowledgments
This work was supported by a Korea Science and Engineering Foundation (KOSEF) grant from the Korean government (MOST) (R13-2002-054-04002-0 and M1075502001-07N5502-00110), and a grant (code #20070301034007) from the BioGreen 21 program, Rural Development Administration, Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Additional information
H.-C. Kim and K.-C. Choi contributed equally to this work.
Electronic supplementary material
Below are the links to the electronic supplementary material.
Supplementary Fig. 1 a
HEK293 cells were transfected with siRNAs or FLAG-tagged CREB3 plasmid. The mRNA and protein levels were determined by qRT-PCR and western blot analysis. b Confirmation of siRNA efficiencies by western blotting and RT-PCR analysis. si-RNAs were transfected into HEK 293 cells. Two days after transfection, the cells were harvested. mRNAs and cell lysates were analysed by western blotting (upper panel) and RT-PCR (lower panel) (TIFF 606 KB)
Supplementary Fig. 2
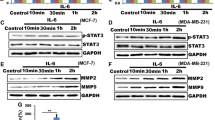
Effect of HDAC3 on the expression of the CCR7 gene. MDA-MB-231 cells were transfected with siHDAC3 or FLAG-tagged CREB3 plasmid. The mRNA levels were determined by qRT-PCR (TIFF 225 kb)
Supplementary Fig. 3
In vitro acetylation assays were performed with HeLa nuclear extract as an enzyme source using GST or GST-fused proteins, and subsequently processed for autoradiography (TIFF 1.50 mb)
Supplementary Fig. 4
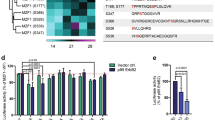
Oncomine 4.3 analysis for the expression patterns of CREB3 in normal and breast cancer tissues (TIFF 675 mb)
Rights and permissions
About this article
Cite this article
Kim, HC., Choi, KC., Choi, HK. et al. HDAC3 selectively represses CREB3-mediated transcription and migration of metastatic breast cancer cells. Cell. Mol. Life Sci. 67, 3499–3510 (2010). https://doi.org/10.1007/s00018-010-0388-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-010-0388-5