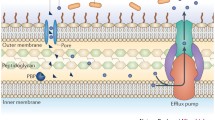
Abstract
Resistance to tetracycline emerged soon after its discovery six decades ago. Extensive clinical and non-clinical uses of this class of antibiotic over the years have combined to select for a large number of resistant determinants, collectively termed the tetracycline resistome. In order to impart resistance, microbes use different molecular mechanisms including target protection, active efflux, and enzymatic degradation. A deeper understanding of the structure, mechanism, and regulation of the genes and proteins associated with tetracycline resistance will contribute to the development of tetracycline derivatives that overcome resistance. Newer generations of tetracyclines derived from engineering of biosynthetic genetic programs, semi-synthesis, and in particular recent developments in their chemical synthesis, together with a growing understanding of resistance, will serve to retain this class of antibiotic to combat pathogens.









Similar content being viewed by others
References
Fluit AC, Florijn A, Verhoef J, Milatovic D (2005) Presence of tetracycline resistance determinants and susceptibility to tigecycline and minocycline. Antimicrob Agents Chemother 49:1636–1638
Charest MG, Siegel DR, Myers AG (2005) Synthesis of minus-tetracycline. J Am Chem Soc 127:8292–8293
Charest MG, Lerner CD, Brubaker JD, Siegel DR, Myers AG (2005) A convergent enantioselective route to structurally diverse 6-deoxytetracycline antibiotics. Science 308:395–398
Lokeshwar BL, Selzer MG, Zhu BQ, Block NL, Golub LM (2002) Inhibition of cell proliferation, invasion, tumor growth and metastasis by an oral non-antimicrobial tetracycline analog (COL-3) in a metastatic prostate cancer model. Int J Cancer 98:297–309
Dezube BJ, Krown SE, Lee JY, Bauer KS, Aboulafia DM (2006) Randomized phase II trial of matrix metalloproteinase inhibitor COL-3 in AIDS-related Kaposi’s sarcoma: an AIDS Malignancy Consortium Study. J Clin Oncol 24:1389–1394
Anokhina MM, Barta A, Nierhaus KH, Spiridonova VA, Kopylov AM (2004) Mapping of the second tetracycline binding site on the ribosomal small subunit of E. coli. Nucleic Acids Res 32:2594–2597
Pioletti M, Schlünzen F, Harms J, Zarivach R, Glühmann M, Avila H, Bashan A, Bartels H, Auerbach T, Jacobi C, Hartsch T, Yonath A, Franceschi F (2001) Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J 20:1829–1839
Brodersen DE, Clemons WM Jr, Carter AP, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V (2000) The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell 103:1143–1154
Aleksandrov A, Simonson T (2008) Molecular dynamics simulations of the 30S ribosomal subunit reveal a preferred tetracycline binding site. J Am Chem Soc 130:1114–1115
Schnappinger D, Hillen W (1996) Tetracyclines: antibiotic action, uptake, and resistance mechanisms. Arch Microbiol 165:359–369
Wright GD (2007) The antibiotic resistome: the nexus of chemical and genetic diversity. Nat Rev Microbiol 5:175–186
D’Costa VM, Griffiths E, Wright GD (2007) Expanding the soil antibiotic resistome: exploring environmental diversity. Curr Opin Microbiol 10:481–489
Davies J, Wright GD (1997) Bacterial resistance to aminoglycoside antibiotics. Trends Microbiol 5:234–240
Liou GF, Yoshizawa S, Courvalin P, Galimand M (2006) Aminoglycoside resistance by ArmA-mediated ribosomal 16S methylation in human bacterial pathogens. J Mol Biol 359:358–364
Marshall CG, Lessard IA, Park I, Wright GD (1998) Glycopeptide antibiotic resistance genes in glycopeptide-producing organisms. Antimicrob Agents Chemother 42:2215–2220
Canton R, Coque TM (2006) The CTX-M beta-lactamase pandemic. Curr Opin Microbiol 9:466–475
Roberts MC (1996) Tetracycline resistance determinants: mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiol Rev 19:1–24
Chopra I, Roberts M (2001) Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev 65:232–260 (second page, table of contents)
Roberts MC (2005) Update on acquired tetracycline resistance genes. FEMS Microbiol Lett 245:195–203
Guillaume G, Ledent V, Moens W, Collard JM (2004) Phylogeny of efflux-mediated tetracycline resistance genes and related proteins revisited. Microb Drug Resist 10:11–26
Agerso Y, Guardabassi L (2005) Identification of Tet 39, a novel class of tetracycline resistance determinant in Acinetobacter spp. of environmental and clinical origin. J Antimicrob Chemother 55:566–569
Thompson SA, Maani EV, Lindell AH, King CJ, McArthur JV (2007) Novel tetracycline resistance determinant isolated from an environmental strain of Serratia marcescens. Appl Environ Microbiol 73:2199–2206
Brown MG, Mitchell EH, Balkwill DL (2008) Tet 42, a novel tetracycline resistance determinant isolated from deep terrestrial subsurface bacteria. Antimicrob Agents Chemother 52:4518–4521
Kazimierczak KA, Rincon MT, Patterson AJ, Martin JC, Young P, Flint HJ, Scott KP (2008) A new tetracycline efflux gene, tet(40), is located in tandem with tet(O/32/O) in a human gut firmicute bacterium and in metagenomic library clones. Antimicrob Agents Chemother 52:4001–4009
Truong-Bolduc QC, Dunman PM, Strahilevitz J, Projan SJ, Hooper DC (2005) MgrA is a multiple regulator of two new efflux pumps in Staphylococcus aureus. J Bacteriol 187:2395–2405
Paulsen IT, Brown MH, Skurray RA (1996) Proton-dependent multidrug efflux systems. Microbiol Rev 60:575–608
Tamura N, Konishi S, Yamaguchi A (2003) Mechanisms of drug/H+ antiport: complete cysteine-scanning mutagenesis and the protein engineering approach. Curr Opin Chem Biol 7:570–579
Yamaguchi A, Someya Y, Sawai T (1992) Metal-tetracycline/H+ antiporter of Escherichia coli encoded by transposon Tn10. The role of a conserved sequence motif, GXXXXRXGRR, in a putative cytoplasmic loop between helices 2 and 3. J Biol Chem 267:19155–19162
Yamaguchi A, Akasaka T, Ono N, Someya Y, Nakatani M, Sawai T (1992) Metal-tetracycline/H+ antiporter of Escherichia coli encoded by transposon Tn10. Roles of the aspartyl residues located in the putative transmembrane helices. J Biol Chem 267:7490–7498
Yamaguchi A, Samejima T, Kimura T, Sawai T (1996) His257 is a uniquely important histidine residue for tetracycline/H+ antiport function but not mandatory for full activity of the transposon Tn10-encoded metal-tetracycline/H+ antiporter. Biochemistry 35:4359–4364
Hillen W, Berens C (1994) Mechanisms underlying expression of Tn10 encoded tetracycline resistance. Annu Rev Microbiol 48:345–369
Saenger W, Orth P, Kisker C, Hillen W, Hinrichs W (2000) The tetracycline repressor-A paradigm for a biological switch. Angew Chem Int Ed Engl 39:2042–2052
Pang Y, Brown BA, Steingrube VA, Wallace RJ Jr, Roberts MC (1994) Tetracycline resistance determinants in mycobacterium and streptomyces species. Antimicrob Agents Chemother 38:1408–1412
Taylor DE, Chau A (1996) Tetracycline resistance mediated by ribosomal protection. Antimicrob Agents Chemother 40:1–5
Kobayashi T, Nonaka L, Maruyama F, Suzuki S (2007) Molecular evidence for the ancient origin of the ribosomal protection protein that mediates tetracycline resistance in bacteria. J Mol Evol 65:228–235
Burdett V (1996) Tet(M)-promoted release of tetracycline from ribosomes is GTP dependent. J Bacteriol 178:3246–3251
Connell SR, Tracz DM, Nierhaus KH, Taylor DE (2003) Ribosomal protection proteins and their mechanism of tetracycline resistance. Antimicrob Agents Chemother 47:3675–3681
Rasmussen BA, Gluzman Y, Tally FP (1994) Inhibition of protein synthesis occurring on tetracycline-resistant, TetM-protected ribosomes by a novel class of tetracyclines, the glycylcyclines. Antimicrob Agents Chemother 38:1658–1660
Melville CM, Scott KP, Mercer DK, Flint HJ (2001) Novel tetracycline resistance gene, tet(32), in the Clostridium-related human colonic anaerobe K10 and its transmission in vitro to the rumen anaerobe Butyrivibrio fibrisolvens. Antimicrob Agents Chemother 45:3246–3249
Patterson AJ, Rincon MT, Flint HJ, Scott KP (2007) Mosaic tetracycline resistance genes are widespread in human and animal fecal samples. Antimicrob Agents Chemother 51:1115–1118
Warburton P, Roberts AP, Allan E, Seville L, Lancaster H, Mullany P (2009) Characterization of tet(32) genes from the oral metagenome. Antimicrob Agents Chemother 53:273–276
Oggioni MR, Dowson CG, Smith JM, Provvedi R, Pozzi G (1996) The tetracycline resistance gene tet(M) exhibits mosaic structure. Plasmid 35:156–163
Stanton TB, McDowall JS, Rasmussen MA (2004) Diverse tetracycline resistance genotypes of Megasphaera elsdenii strains selectively cultured from swine feces. Appl Environ Microbiol 70:3754–3757
van Hoek AH, Mayrhofer S, Domig KJ, Florez AB, Ammor MS, Mayo B, Aarts HJ (2008) Mosaic tetracycline resistance genes and their flanking regions in Bifidobacterium thermophilum and Lactobacillus johnsonii. Antimicrob Agents Chemother 52:248–252
Speer BS, Salyers AA (1989) Novel aerobic tetracycline resistance gene that chemically modifies tetracycline. J Bacteriol 171:148–153
Yang W, Moore IF, Koteva KP, Bareich DC, Hughes DW, Wright GD (2004) TetX is a flavin-dependent monooxygenase conferring resistance to tetracycline antibiotics. J Biol Chem 279:52346–52352
Moore IF, Hughes DW, Wright GD (2005) Tigecycline is modified by the flavin-dependent monooxygenase TetX. Biochemistry 44:11829–11835
Ghosh S, Sadowsky MJ, Roberts MC, Gralnick JA, LaPara TM (2009) Sphingobacterium sp. strain PM2–P1-29 harbours a functional tet(X) gene encoding for the degradation of tetracycline. J Appl Microbiol 106:1336–1342
Nonaka L, Suzuki S (2002) New Mg2+-dependent oxytetracycline resistance determinant tet 34 in Vibrio isolates from marine fish intestinal contents. Antimicrob Agents Chemother 46:1550–1552
Diaz-Torres ML, McNab R, Spratt DA, Villedieu A, Hunt N, Wilson M, Mullany P (2003) Novel tetracycline resistance determinant from the oral metagenome. Antimicrob Agents Chemother 47:1430–1432
Petkovic H, Cullum J, Hranueli D, Hunter IS, Perić-Concha N, Pigac J, Thamchaipenet A, Vujaklija D, Long PF (2006) Genetics of Streptomyces rimosus, the oxytetracycline producer. Microbiol Mol Biol Rev 70:704–728
Ohnuki T, Katoh T, Imanaka T, Aiba S (1985) Molecular cloning of tetracycline resistance genes from Streptomyces rimosus in Streptomyces griseus and characterization of the cloned genes. J Bacteriol 161:1010–1016
Dittrich W, Schrempf H (1992) The unstable tetracycline resistance gene of Streptomyces lividans 1326 encodes a putative protein with similarities to translational elongation factors and Tet(M) and Tet(O) proteins. Antimicrob Agents Chemother 36:1119–1124
McMurry LM, Levy SB (1998) Revised sequence of OtrB (tet347) tetracycline efflux protein from Streptomyces rimosus. Antimicrob Agents Chemother 42:3050
Cohen SP, McMurry LM, Levy SB (1988) marA locus causes decreased expression of OmpF porin in multiple-antibiotic-resistant (Mar) mutants of Escherichia coli. J Bacteriol 170:5416–5422
Eady EA, Jones CE, Gardner KJ, Taylor JP, Cove JH, Cunliffe WJ (1993) Tetracycline-resistant propionibacteria from acne patients are cross-resistant to doxycycline, but sensitive to minocycline. Br J Dermatol 128:556–560
Kurokawa I, Nishijima S, Asada Y (1988) The antibiotic susceptibility of Propionibacterium acnes: a 15-year bacteriological study and retrospective evaluation. J Dermatol 15:149–154
Leyden JJ, McGinley KJ, Cavalieri S, Webster GF, Mills OH, Kligman AM (1983) Propionibacterium acnes resistance to antibiotics in acne patients. J Am Acad Dermatol 8:41–45
Ross JI, Eady EA, Cove JH, Cunliffe WJ (1998) 16S rRNA mutation associated with tetracycline resistance in a gram-positive bacterium. Antimicrob Agents Chemother 42:1702–1705
Gerrits MM, de Zoete MR, Arents NL, Kuipers EJ, Kusters JG (2002) 16S rRNA mutation-mediated tetracycline resistance in Helicobacter pylori. Antimicrob Agents Chemother 46:2996–3000
Trieber CA, Taylor DE (2002) Mutations in the 16S rRNA genes of Helicobacter pylori mediate resistance to tetracycline. J Bacteriol 184:2131–2140
Dailidiene D, Bertoli MT, Miciuleviciene J, Mukhopadhyay AK, Dailide G, Pascasio MA, Kupcinskas L, Berg DE (2002) Emergence of tetracycline resistance in Helicobacter pylori: multiple mutational changes in 16S ribosomal DNA and other genetic loci. Antimicrob Agents Chemother 46:3940–3946
Gerrits MM, Berning M, Van Vliet AH, Kuipers EJ, Kusters JG (2003) Effects of 16S rRNA gene mutations on tetracycline resistance in Helicobacter pylori. Antimicrob Agents Chemother 47:2984–2986
Levy SB, McMurry LM, Burdett V, Courvalin P, Hillen W, Roberts MC, Taylor DE (1989) Nomenclature for tetracycline resistance determinants. Antimicrob Agents Chemother 33:1373–1374
Levy SB, McMurry LM, Roberts MC (2005) Tet protein hybrids. Antimicrob Agents Chemother 49:3099
Liu B, Pop M (2009) ARDB—antibiotic resistance genes database. Nucleic Acids Res 37:D443–D447
D’Costa VM, McGrann KM, Hughes DW, Wright GD (2006) Sampling the antibiotic resistome. Science 311:374–377
Baltz RH (2007) Antimicrobials from actinomycetes: back to the future. Microbe 2:125–131
Hinrichs W, Kisker C, Duvel M, Muller A, Tovar K, Hillen W, Saenger W (1994) Structure of the Tet repressor–tetracycline complex and regulation of antibiotic resistance. Science 264:418–420
Chopra I (1994) Tetracycline analogs whose primary target is not the bacterial ribosome. Antimicrob Agents Chemother 38:637–640
Sun C, Wang Q, Brubaker JD, Wright PM, Lerner CD, Noson K, Carest M, Siegel DR, Wang YM, Myers AG (2008) A robust platform for the synthesis of new tetracycline antibiotics. J Am Chem Soc 130:17913–17927
Levy SB, McMurry LM, Brdett V, Courvalin P, Hillen W, Roberts MC, Taylor DE (1999) Nomenclature for new tetracycline resistance determinants. Antimicrob Agents Chemother 43:1523–1524
Acknowledgments
This work is supported by the Natural Sciences and Engineering Research Council of Canada and the Canadian Institutes of Health Research.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Thaker, M., Spanogiannopoulos, P. & Wright, G.D. The tetracycline resistome. Cell. Mol. Life Sci. 67, 419–431 (2010). https://doi.org/10.1007/s00018-009-0172-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-009-0172-6