Abstract
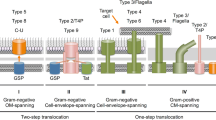
In the three domains of life, the Sec, YidC/Oxa1, and Tat translocases play important roles in protein translocation across membranes and membrane protein insertion. While extensive studies have been performed on the endoplasmic reticular and Escherichia coli systems, far fewer studies have been done on archaea, other Gram-negative bacteria, and Gram-positive bacteria. Interestingly, work carried out to date has shown that there are differences in the protein transport systems in terms of the number of translocase components and, in some cases, the translocation mechanisms and energy sources that drive translocation. In this review, we will describe the different systems employed to translocate and insert proteins across or into the cytoplasmic membrane of archaea and bacteria.





Similar content being viewed by others
References
Driessen AJ, Nouwen N (2008) Protein translocation across the bacterial cytoplasmic membrane. Annu Rev Biochem 77:643–667
Luirink J, ten Hagen-Jongman CM, van der Weijden CC, Oudega B, High S, Dobberstein B, Kusters R (1994) An alternative protein targeting pathway in Escherichia coli: studies on the role of FtsY. EMBO J 13:2289–2296
Mainprize IL, Vulcu F, Andrews DW (2007) The signal recognition particle and its receptor in the ER protein targeting in the enzymes vol 25, 3rd edn. In: Dalbey RE, Koehler CM, Tamanoi F (eds) Molecular machines involved in protein transport across cellular membranes. Academic Press, San Diego, pp 177–206
Alken M, Hegde R (2007) The translocation apparatus of the endoplasmic reticulum in the enzymes, vol 25, 3rd edn. In: Dalbey RE, Koehler CM, Tamanoi F (eds) Molecular machines involved in protein transport across cellular membranes. Academic Press, San Diego, pp 207–243
Nakatsukasa K, Brodsky JL (2007) The role of Bip/Kar2p in the translocation of proteins across the ER membrane in the enzymes, vol 25, 3rd edn. In: Dalbey RE, Koehler CM, Tamanoi F (eds) Molecular machines involved in protein transport across cellular membranes. Academic Press, San Diego, pp 245–273
von Heijne G (1999) Recent advances in the understanding of membrane protein assembly and structure. Q Rev Biophys 32:285–307
von Heijne G (1986) The distribution of positively charged residues in bacterial inner membrane proteins correlates with the trans-membrane topology. EMBO J 5:3021–3027
von Heijne G, Gavel Y (1988) Topogenic signals in integral membrane proteins. Eur J Biochem 174:671–678
Gavel Y, von Heijne G (1992) The distribution of charged amino acids in mitochondrial inner-membrane proteins suggests different modes of membrane integration for nuclearly and mitochondrially encoded proteins. Eur J Biochem 205:1207–1215
Dalbey RE (1990) Positively charged residues are important determinants of membrane protein topology. Trends Biochem Sci 15:253–257
Ruiz N, Kahne D, Silhavy TJ (2006) Advances in understanding bacterial outer-membrane biogenesis. Nat Rev Microbiol 4:57–66
von Heijne G (1981) Membrane proteins: the amino acid composition of membrane-penetrating segments. Eur J Biochem 120:275–278
Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105–132
Hopp TP, Woods KR (1981) Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci USA 78:3824–3828
Eisenberg D, Weiss RM, Terwilliger TC (1982) The helical hydrophobic moment: a measure of the amphiphilicity of a helix. Nature 299:371–374
von Heijne G (1992) Membrane protein structure prediction Hydrophobicity analysis and the positive-inside rule. J Mol Biol 225:487–494
Tusnady GE, Simon I (1998) Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J Mol Biol 283:489–506
Sonnhammer EL, von Heijne G, Krogh A (1998) A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol 6:175–182
Persson B, Argos P (1997) Prediction of membrane protein topology utilizing multiple sequence alignments. J Protein Chem 16:453–457
Jones DT, Taylor WR, Thornton JM (1994) A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry 33:3038–3049
Rost B, Casadio R, Fariselli P, Sander C (1995) Transmembrane helices predicted at 95% accuracy. Protein Sci 4:521–533
Moller S, Croning MD, Apweiler R (2001) Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics 17:646–653
Bernsel A, Viklund H, Falk J, Lindahl E, von Heijne G, Elofsson A (2008) Prediction of membrane-protein topology from first principles. Proc Natl Acad Sci USA 105:7177–7181
Viklund H, Elofsson A (2004) Best alpha-helical transmembrane protein topology predictions are achieved using hidden Markov models and evolutionary information. Protein Sci 13:1908–1917
Jones DT (1998) Do transmembrane protein superfolds exist? FEBS Lett 423:281–285
Jones DT (2007) Improving the accuracy of transmembrane protein topology prediction using evolutionary information. Bioinformatics 23:538–544
Kall L, Krogh A, Sonnhammer EL (2004) A combined transmembrane topology and signal peptide prediction method. J Mol Biol 338:1027–1036
Kall L, Krogh A, Sonnhammer EL (2005) An HMM posterior decoder for sequence feature prediction that includes homology information. Bioinformatics 21[Suppl 1]:i251–i257
Viklund H, Elofsson A (2008) OCTOPUS: improving topology prediction by two-track ANN-based preference scores and an extended topological grammar. Bioinformatics 24:1662–1668
Bernsel A, Viklund H, Hennerdal A, Elofsson A (2009) TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Res 37[Suppl 2]:W465–W468. doi: 10.1093/nar/gkp363
Keenan RJ, Freymann DM, Stroud RM, Walter P (2001) The signal recognition particle. Annu Rev Biochem 70:755–775
Walter P, Blobel G (1981) Translocation of proteins across the endoplasmic reticulum III Signal recognition protein (SRP) causes signal sequence-dependent and site- specific arrest of chain elongation that is released by microsomal membranes. J Cell Biol 91:557–561
Tajima S, Lauffer L, Rath VL, Walter P (1986) The signal recognition particle receptor is a complex that contains two distinct polypeptide chains. J Cell Biol 103:1167–1178
Valent QA, Scotti PA, High S, de Gier JW, von Heijne G, Lentzen G, Wintermeyer W, Oudega B, Luirink J (1998) The Escherichia coli SRP and SecB targeting pathways converge at the translocon. EMBO J 17:2504–2512
Prinz A, Behrens C, Rapoport TA, Hartmann E, Kalies KU (2000) Evolutionarily conserved binding of ribosomes to the translocation channel via the large ribosomal RNA. EMBO J 19:1900–1906
Bradshaw N, Neher SB, Booth DS, Walter P (2009) Signal sequences activate the catalytic switch of SRP RNA. Science 323:127–130
Ulbrandt ND, Newitt JA, Bernstein HD (1997) The E. coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell 88:187–196
Luirink J, Sinning I (2004) SRP-mediated protein targeting: structure and function revisited. Biochim Biophys Acta 1694:17–35
Phillips GJ, Silhavy TJ (1992) The E. coli ffh gene is necessary for viability and efficient protein export. Nature 359:744–746
Zanen G, Antelmann H, Meima R, Jongbloed JD, Kolkman M, Hecker M, van Dijl JM, Quax WJ (2006) Proteomic dissection of potential signal recognition particle dependence in protein secretion by Bacillus subtilis. Proteomics 6:3636–3648
Zanen G, Houben EN, Meima R, Tjalsma H, Jongbloed JD, Westers H, Oudega B, Luirink J, van Dijl JM, Quax WJ (2005) Signal peptide hydrophobicity is critical for early stages in protein export by Bacillus subtilis. FEBS J 272:4617–4630
Nakamura K, Nishiguchi M, Honda K, Yamane K (1994) The Bacillus subtilis SRP54 homologue, Ffh, has an intrinsic GTPase activity and forms a ribonucleoprotein complex with small cytoplasmic RNA in vivo. Biochem Biophys Res Commun 199:1394–1399
Oguro A, Kakeshita H, Honda K, Takamatsu H, Nakamura K, Yamane K (1995) srb: a Bacillus subtilis gene encoding a homologue of the alpha-subunit of the mammalian signal recognition particle receptor. DNA Res 2:95–100
Nakamura K, Yahagi S, Yamazaki T, Yamane K (1999) Bacillus subtilis histone-like protein, HBsu, is an integral component of a SRP-like particle that can bind the Alu domain of small cytoplasmic RNA. J Biol Chem 274:13569–13576
Struck JC, Vogel DW, Ulbrich N, Erdmann VA (1988) The Bacillus subtilis scRNA is related to the 4.5S RNA from Escherichia coli. Nucleic Acids Res 16:2719
Nishiguchi M, Honda K, Amikura R, Nakamura K, Yamane K (1994) Structural requirements of Bacillus subtilis small cytoplasmic RNA for cell growth, sporulation, and extracellular enzyme production. J Bacteriol 176:157–165
Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, Boland F, Brignell SC, Bron S, Bunai K, Chapuis J, Christiansen LC, Danchin A, Debarbouille M, Dervyn E, Deuerling E, Devine K, Devine SK, Dreesen O, Errington J, Fillinger S, Foster SJ, Fujita Y, Galizzi A, Gardan R, Eschevins C, Fukushima T, Haga K, Harwood CR, Hecker M, Hosoya D, Hullo MF, Kakeshita H, Karamata D, Kasahara Y, Kawamura F, Koga K, Koski P, Kuwana R, Imamura D, Ishimaru M, Ishikawa S, Ishio I, Le Coq D, Masson A, Mauel C, Meima R, Mellado RP, Moir A, Moriya S, Nagakawa E, Nanamiya H, Nakai S, Nygaard P, Ogura M, Ohanan T, O’Reilly M, O’Rourke M, Pragai Z, Pooley HM, Rapoport G, Rawlins JP, Rivas LA, Rivolta C, Sadaie A, Sadaie Y, Sarvas M, Sato T, Saxild HH, Scanlan E, Schumann W, Seegers JF, Sekiguchi J, Sekowska A, Seror SJ, Simon M, Stragier P, Studer R, Takamatsu H, Tanaka T, Takeuchi M, Thomaides HB, Vagner V, van Dijl JM, Watabe K, Wipat A, Yamamoto H, Yamamoto M, Yamamoto Y, Yamane K, Yata K, Yoshida K, Yoshikawa H, Zuber U, Ogasawara N (2003) Essential Bacillus subtilis genes. Proc Natl Acad Sci USA 100:4678–4683
Nakamura K, Imai Y, Nakamura A, Yamane K (1992) Small cytoplasmic RNA of Bacillus subtilis: functional relationship with human signal recognition particle 7S RNA and Escherichia coli 4.5S RNA. J Bacteriol 174:2185–2192
Forsyth RA, Haselbeck RJ, Ohlsen KL, Yamamoto RT, Xu H, Trawick JD, Wall D, Wang L, Brown-Driver V, Froelich JM, King CKGP, McCarthy M, Malone C, Misiner B, Robbins D, Tan Z, Zhu ZY,Carr G, Mosca DA, Zamudio C, Foulkes JG, Zyskind, JW (2002) A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol Microbiol 43:1387–1400
Thanassi JA, Hartman-Neumann SL, Dougherty TJ, Dougherty BA, Pucci MJ (2002) Identification of 113 conserved essential genes using a high-throughput gene disruption system in Streptococcus pneumoniae. Nucleic Acids Res 30:3152–3162
Carpenter PB, Hanlon DW, Ordal GW (1992) flhF, a Bacillus subtilis flagellar gene that encodes a putative GTP-binding protein. Mol Microbiol 6:2705–2713
Salvetti S, Ghelardi E, Celandroni F, Ceragioli M, Giannessi F, Senesi S (2007) FlhF, a signal recognition particle-like GTPase, is involved in the regulation of flagellar arrangement, motility behaviour and protein secretion in Bacillus cereus. Microbiology 153:2541–2552
Bange G, Petzold G, Wild K, Parlitz RO, Sinning I (2007) The crystal structure of the third signal-recognition particle GTPase FlhF reveals a homodimer with bound. GTP Proc Natl Acad Sci USA 104:13621–13625
Zanen G, Antelmann H, Westers H, Hecker M, van Dijl JM, Quax WJ (2004) FlhF, the third signal recognition particle-GTPase of Bacillus subtilis, is dispensable for protein secretion. J Bacteriol 186:5956–5960
Crowley PJ, Svensater G, Snoep JL, Bleiweis AS, Brady LJ (2004) An ffh mutant of Streptococcus mutans is viable and able to physiologically adapt to low pH in continuous culture. FEMS Microbiol Lett 234:315–324
Hasona A, Crowley PJ, Levesque CM, Mair RW, Cvitkovitch DG, Bleiweis AS, Brady LJ (2005) Streptococcal viability and diminished stress tolerance in mutants lacking the signal recognition particle pathway or YidC2. Proc Natl Acad Sci USA 102:17466–17471
Kremer BH, van der Kraan M, Crowley PJ, Hamilton IR, Brady LJ, Bleiweis AS (2001) Characterization of the sat operon in Streptococcus mutans: evidence for a role of Ffh in acid tolerance. J Bacteriol 183:2543–2552
Hasona A, Zuobi-Hasona K, Crowley PJ, Abranches J, Ruelf MA, Bleiweis AS, Brady LJ (2007) Membrane composition changes and physiological adaptation by Streptococcus mutans signal recognition particle pathway mutants. J Bacteriol 189:1219–1230
Rose RW, Bruser T, Kissinger JC, Pohlschroder M (2002) Adaptation of protein secretion to extremely high-salt conditions by extensive use of the twin-arginine translocation pathway. Mol Microbiol 45:943–950
Zwieb C, Eichler J (2002) Getting on target: the archaeal signal recognition particle. Archaea 1:27–34
Kaine BP (1990) Structure of the archaebacterial 7S RNA molecule. Mol Gen Genet 221:315–321
Hainzl T, Huang S, Sauer-Eriksson AE (2007) Interaction of signal-recognition particle 54 GTPase domain and signal-recognition particle RNA in the free signal-recognition particle. Proc Natl Acad Sci USA 104:14911–14916
Bernstein HD, Poritz MA, Strub K, Hoben PJ, Brenner S, Walter P (1989) Model for signal sequence recognition from amino-acid sequence of 54 K subunit of signal recognition particle. Nature 340:482–486
Romisch K, Webb J, Herz J, Prehn S, Frank R, Vingron M, Dobberstein B (1989) Homology of 54 K protein of signal-recognition particle, docking protein and two E. coli proteins with putative GTP-binding domains. Nature 340:478–482
Egea PF, Shan SO, Napetschnig J, Savage DF, Walter P, Stroud RM (2004) Substrate twinning activates the signal recognition particle and its receptor. Nature 427:215–221
Focia PJ, Shepotinovskaya IV, Seidler JA, Freymann DM (2004) Heterodimeric GTPase core of the SRP targeting complex. Science 303:373–377
Bhuiyan SH, Gowda K, Hotokezaka H, Zwieb C (2000) Assembly of archaeal signal recognition particle from recombinant components. Nucleic Acids Res 28:1365–1373
Diener JL, Wilson C (2000) Role of SRP19 in assembly of the Archaeoglobus fulgidus signal recognition particle. Biochemistry 39:12862–12874
Tozik I, Huang Q, Zwieb C, Eichler J (2002) Reconstitution of the signal recognition particle of the halophilic archaeon Haloferax volcanii. Nucleic Acids Res 30:4166–4175
Hainzl T, Huang S, Sauer-Eriksson AE (2005) Structural insights into SRP RNA: an induced fit mechanism for SRP assembly. RNA 11:1043–1050
Moll R, Schmidtke S, Schafer G (1999) Domain structure, GTP-hydrolyzing activity and 7S RNA binding of Acidianus ambivalens ffh-homologous protein suggest an SRP-like complex in archaea. Eur J Biochem 259:441–448
Yurist S, Dahan I, Eichler J (2007) SRP19 is a dispensable component of the signal recognition particle in Archaea. J Bacteriol 189:276–279
Egea PF, Napetschnig J, Walter P, Stroud RM (2008) Structures of SRP54 and SRP19, the two proteins that organize the ribonucleic core of the signal recognition particle from Pyrococcus furiosus. PLoS ONE 3:e3528
Walter P, Johnson AE (1994) Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu Rev Cell Biol 10:87–119
Lutcke H (1995) Signal recognition particle (SRP) a ubiquitous initiator of protein translocation. Eur J Biochem 228:531–550
Stroud RM, Walter P (1999) Signal sequence recognition and protein targeting. Curr Opin Struct Biol 9:754–759
Chang DY, Newitt JA, Hsu K, Bernstein HD, Maraia RJ (1997) A highly conserved nucleotide in the Alu domain of SRP RNA mediates translation arrest through high affinity binding to SRP9/14. Nucleic Acids Res 25:1117–1122
Zwieb C, Samuelsson T (2000) SRPDB (signal recognition particle database). Nucleic Acids Res 28:171–172
de Leeuw E, Poland D, Mol O, Sinning I, ten Hagen-Jongman CM, Oudega B, Luirink J (1997) Membrane association of FtsY, the E. coli SRP receptor. FEBS Lett 416:225–229
Powers T, Walter P (1997) Co-translational protein targeting catalyzed by the Escherichia coli signal recognition particle and its receptor. EMBO J 16:4880–4886
Zelazny A, Seluanov A, Cooper A, Bibi E (1997) The NG domain of the prokaryotic signal recognition particle receptor, FtsY, is fully functional when fused to an unrelated integral membrane polypeptide. Proc Natl Acad Sci USA 94:6025–6029
Egea PF, Tsuruta H, de Leon GP, Napetschnig J, Walter P, Stroud RM (2008) Structures of the signal recognition particle receptor from the archaeon Pyrococcus furiosus: implications for the targeting step at the membrane. PLoS ONE 3:e3619
Gawronski-Salerno J, Coon JST, Focia PJ, Freymann DM (2007) X-ray structure of the T. aquaticus FtsY:GDP complex suggests functional roles for the C-terminal helix of the SRP GTPases. Proteins 66:984–995
Moll R, Schmidtke S, Schaefer G (1996) A putative signal recognition particle receptor alpha subunit (SR alpha) homologue is expressed in the hyperthermophilic crenarchaeon Sulfolobus acidocaldarius. FEMS Microbiol Lett 137:51–56
Moll R, Schmidtke S, Petersen A, Schafer G (1997) The signal recognition particle receptor alpha subunit of the hyperthermophilic archaeon Acidianus ambivalens exhibits an intrinsic GTP-hydrolyzing activity. Biochim Biophys Acta 1335:218–230
Eichler J (2000) Archaeal protein translocation crossing membranes in the third domain of life. Eur J Biochem 267:3402–3412
Rapoport TA (2007) Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 450:663–669
Vrontou E, Economou A (2004) Structure and function of SecA, the preprotein translocase nanomotor. Biochim Biophys Acta 1694:67–80
Deitermann S, Sprie GS, Koch HG (2005) A dual function for SecA in the assembly of single spanning membrane proteins in Escherichia coli. J Biol Chem 280:39077–39085
Economou A, Wickner W (1994) SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell 78:835–843
Schiebel E, Driessen AJ, Hartl FU, Wickner W (1991) Delta mu H+ and ATP function at different steps of the catalytic cycle of preprotein translocase. Cell 64:927–939
van der Wolk JP, de Wit JG, Driessen AJ (1997) The catalytic cycle of the Escherichia coli SecA ATPase comprises two distinct preprotein translocation events. EMBO J 16:7297–7304
Pogliano JA, Beckwith J (1994) SecD and SecF facilitate protein export in Escherichia coli. EMBO J 13:554–561
Chen M, Xie K, Yuan J, Yi L, Facey SJ, Pradel N, Wu LF, Kuhn A, Dalbey RE (2005) Involvement of SecDF and YidC in the membrane insertion of M13 procoat mutants. Biochemistry 44:10741–10749
Kiefer D, Kuhn A (2007) YidC as an essential and multifunctional component in membrane protein assembly. Int Rev Cytol 259:113–138
Van Der Laan M, Urbanus ML, Ten Hagen-Jongman CM, Nouwen N, Oudega B, Harms N, Driessen AJ, Luirink J (2003) A conserved function of YidC in the biogenesis of respiratory chain complexes. Proc Natl Acad Sci USA 100:5801–5806
Price CE, Driessen AJ (2008) YidC is involved in the biogenesis of anaerobic respiratory complexes in the inner membrane of Escherichia coli. J Biol Chem 283:26921–26927
Samuelson JC, Chen M, Jiang F, Moller I, Wiedmann M, Kuhn A, Phillips GJ, Dalbey RE (2000) YidC mediates membrane protein insertion in bacteria. Nature 406:637–641
Jungnickel B, Rapoport TA, Hartmann E (1994) Protein translocation: common themes from bacteria to man. FEBS Lett 346:73–77
Lyman SK, Schekman R (1997) Binding of secretory precursor polypeptides to a translocon subcomplex is regulated by BiP. Cell 88:85–96
Sanders SL, Whitfield KM, Vogel JP, Rose MD, Schekman RW (1992) Sec61p and BiP directly facilitate polypeptide translocation into the ER. Cell 69:353–365
von Heijne G, Abrahmsen L (1989) Species-specific variation in signal peptide design Implications for protein secretion in foreign hosts. FEBS Lett 244:439–446
van Wely KH, Swaving J, Broekhuizen CP, Rose M, Quax WJ, Driessen AJ (1999) Functional identification of the product of the Bacillus subtilis yvaL gene as a SecG homologue. J Bacteriol 181:1786–1792
Schatz PJ, Bieker KL, Ottemann KM, Silhavy TJ, Beckwith J (1991) One of three transmembrane stretches is sufficient for the functioning of the SecE protein, a membrane component of the E. coli secretion machinery. EMBO J 10:1749–1757
Swaving J, van Wely KH, Driessen AJ (1999) Preprotein translocation by a hybrid translocase composed of Escherichia coli and Bacillus subtilis subunits. J Bacteriol 181:7021–7027
Rigel NW, Braunstein M (2008) A new twist on an old pathway—accessory Sec [corrected] systems. Mol Microbiol 69:291–302
Sibbald MJ, Ziebandt AK, Engelmann S, Hecker M, de Jong A, Harmsen HJ, Raangs GC, Stokroos I, Arends JP, Dubois JY, van Dijl JM (2006) Mapping the pathways to staphylococcal pathogenesis by comparative secretomics. Microbiol Mol Biol Rev 70:755–788
Caspers M, Freudl R (2008) Corynebacterium glutamicum possesses two secA homologous genes that are essential for viability. Arch Microbiol 189:605–610
Bensing BA, Sullam PM (2002) An accessory sec locus of Streptococcus gordonii is required for export of the surface protein GspB and for normal levels of binding to human platelets. Mol Microbiol 44:1081–1094
Wu H, Bu S, Newell P, Chen Q, Fives-Taylor P (2007) Two gene determinants are differentially involved in the biogenesis of Fap1 precursors in Streptococcus parasanguis. J Bacteriol 189:1390–1398
Takamatsu D, Bensing BA, Sullam PM (2004) Genes in the accessory sec locus of Streptococcus gordonii have three functionally distinct effects on the expression of the platelet-binding protein GspB. Mol Microbiol 52:189–203
Bensing BA, Takamatsu D, Sullam PM (2005) Determinants of the streptococcal surface glycoprotein GspB that facilitate export by the accessory Sec system. Mol Microbiol 58:1468–1481
Chen Q, Wu H, Fives-Taylor PM (2004) Investigating the role of secA2 in secretion and glycosylation of a fimbrial adhesin in Streptococcus parasanguis FW213. Mol Microbiol 53:843–856
Gibbons HS, Wolschendorf F, Abshire M, Niederweis M, Braunstein M (2007) Identification of two Mycobacterium smegmatis lipoproteins exported by a SecA2-dependent pathway. J Bacteriol 189:5090–5100
Bensing BA, Siboo IR, Sullam PM (2007) Glycine residues in the hydrophobic core of the GspB signal sequence route export toward the accessory Sec pathway. J Bacteriol 189:3846–3854
Bensing BA, Sullam PM (2009) Characterization of Streptococcus gordonii SecA2 as a paralogue of SecA. J Bacteriol 191:3482–3491
Braunstein M, Espinosa BJ, Chan J, Belisle JT, Jacobs WR Jr (2003) SecA2 functions in the secretion of superoxide dismutase A and in the virulence of Mycobacterium tuberculosis. Mol Microbiol 48:453–464
Chen Q, Sun B, Wu H, Peng Z, Fives-Taylor PM (2007) Differential roles of individual domains in selection of secretion route of a Streptococcus parasanguinis serine-rich adhesin, Fap1. J Bacteriol 189:7610–7617
Bolhuis A, Broekhuizen CP, Sorokin A, van Roosmalen ML, Venema G, Bron S, Quax WJ, van Dijl JM (1998) SecDF of Bacillus subtilis, a molecular Siamese twin required for the efficient secretion of proteins. J Biol Chem 273:21217–21224
Zweers JC, Barak I, Becher D, Driessen AJ, Hecker M, Kontinen VP, Saller MJ, Vavrova L, van Dijl JM (2008) Towards the development of Bacillus subtilis as a cell factory for membrane proteins and protein complexes. Microb Cell Fact 7:10
Bunai K, Nozaki M, Kakeshita H, Nemoto T, Yamane K (2005) Quantitation of de novo localized (15)N-labeled lipoproteins and membrane proteins having one and two transmembrane segments in a Bacillus subtilis secA temperature-sensitive mutant using 2D-PAGE and MALDI-TOF MS. J Proteome Res 4:826–836
Bunai K, Yamane K (2005) Effectiveness and limitation of two-dimensional gel electrophoresis in bacterial membrane protein proteomics and perspectives. J Chromatogr B Analyt Technol Biomed Life Sci 815:227–236
Dreisbach A, Otto A, Becher D, Hammer E, Teumer A, Gouw JW, Hecker M, Volker U (2008) Monitoring of changes in the membrane proteome during stationary phase adaptation of Bacillus subtilis using in vivo labeling techniques. Proteomics 8:2062–2076
Wolff S, Hahne H, Hecker M, Becher D (2008) Complementary analysis of the vegetative membrane proteome of the human pathogen Staphylococcus aureus. Mol Cell Proteomics 7:1460–1468
Hahne H, Wolff S, Hecker M, Becher D (2008) From complementarity to comprehensiveness—targeting the membrane proteome of growing Bacillus subtilis by divergent approaches. Proteomics 8:4123–4136
Campo N, Tjalsma H, Buist G, Stepniak D, Meijer M, Veenhuis M, Westermann M, Muller JP, Bron S, Kok J, Kuipers OP, Jongbloed JD (2004) Subcellular sites for bacterial protein export. Mol Microbiol 53:1583–1599
Rosch J, Caparon M (2004) A microdomain for protein secretion in Gram-positive bacteria. Science 304:1513–1515
Rosch JW, Caparon MG (2005) The ExPortal: an organelle dedicated to the biogenesis of secreted proteins in Streptococcus pyogenes. Mol Microbiol 58:959–968
Hunt JF, Weinkauf S, Henry L, Fak JJ, McNicholas P, Oliver DB, Deisenhofer J (2002) Nucleotide control of interdomain interactions in the conformational reaction cycle of SecA. Science 297:2018–2026
Sharma V, Arockiasamy A, Ronning DR, Savva CG, Holzenburg A, Braunstein M, Jacobs WR Jr, Sacchettini JC (2003) Crystal structure of Mycobacterium tuberculosis SecA, a preprotein translocating ATPase. Proc Natl Acad Sci USA 100:2243–2248
Osborne AR, Clemons WM Jr, Rapoport TA (2004) A large conformational change of the translocation ATPase SecA. Proc Natl Acad Sci USA 101:10937–10942
Vassylyev DG, Mori H, Vassylyeva MN, Tsukazaki T, Kimura Y, Tahirov TH, Ito K (2006) Crystal structure of the translocation ATPase SecA from Thermus thermophilus reveals a parallel, head-to-head dimer. J Mol Biol 364:248–258
Papanikolau Y, Papadovasilaki M, Ravelli RB, McCarthy AA, Cusack S, Economou A, Petratos K (2007) Structure of dimeric SecA, the Escherichia coli preprotein translocase motor. J Mol Biol 366:1545–1557
Zimmer J, Nam Y, Rapoport TA (2008) Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature 455:936–943
Papanikou E, Karamanou S, Economou A (2007) Bacterial protein secretion through the translocase nanomachine. Nat Rev Microbiol 5:839–851
Cooper DB, Smith VF, Crane JM, Roth HC, Lilly AA, Randall LL (2008) SecA, the motor of the secretion machine, binds diverse partners on one interactive surface. J Mol Biol 382:74–87
Erlandson KJ, Miller SB, Nam Y, Osborne AR, Zimmer J, Rapoport TA (2008) A role for the two-helix finger of the SecA ATPase in protein translocation. Nature 455:984–987
Tsukazaki T, Mori H, Fukai S, Ishitani R, Mori T, Dohmae N, Perederina A, Sugita Y, Vassylyev DG, Ito K, Nureki O (2008) Conformational transition of Sec machinery inferred from bacterial SecYE structures. Nature 455:988–991
Auer J, Spicker G, Bock A (1991) Presence of a gene in the archaebacterium Methanococcus vannielii homologous to secY of eubacteria. Biochimie 73:683–688
Cao TB, Saier MH Jr (2003) The general protein secretory pathway: phylogenetic analyses leading to evolutionary conclusions. Biochim Biophys Acta 1609:115–125
Irihimovitch V, Eichler J (2003) Post-translational secretion of fusion proteins in the halophilic archaea Haloferax volcanii. J Biol Chem 278:12881–12887
Kinch LN, Saier MH Jr, Grishin NV (2002) Sec61beta—a component of the archaeal protein secretory system. Trends Biochem Sci 27:170–171
Van den Berg B, Clemons WM Jr, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA (2004) X-ray structure of a protein-conducting channel. Nature 427:36–44
Plath K, Mothes W, Wilkinson BM, Stirling CJ, Rapoport TA (1998) Signal sequence recognition in posttranslational protein transport across the yeast. ER Membrane Cell 94:795–807
Pohlschroder M, Hartmann E, Hand NJ, Dilks K, Haddad A (2005) Diversity and evolution of protein translocation. Annu Rev Microbiol 59:91–111
Eichler J (2003) Evolution of the prokaryotic protein translocation complex: a comparison of archaeal and bacterial versions of SecDF. Mol Phylogenet Evol 27:504–509
Duong F, Wickner W (1997) The SecDFyajC domain of preprotein translocase controls preprotein movement by regulating SecA membrane cycling. EMBO J 16:4871–4879
Economou A, Pogliano JA, Beckwith J, Oliver DB, Wickner W (1995) SecA membrane cycling at SecYEG is driven by distinct ATP binding and hydrolysis events and is regulated by SecD and SecF. Cell 83:1171–1181
Nouwen N, Piwowarek M, Berrelkamp G, Driessen AJ (2005) The large first periplasmic loop of SecD and SecF plays an important role in SecDF functioning. J Bacteriol 187:5857–5860
Pohlschroder M, Prinz WA, Hartmann E, Beckwith J (1997) Protein translocation in the three domains of life: variations on a theme. Cell 91:563–566
Yi L, Dalbey RE (2005) Oxa1/Alb3/YidC system for insertion of membrane proteins in mitochondria, chloroplasts and bacteria (review). Mol Membr Biol 22:101–111
Henry R, Goforth RL, Schunemann (2007) Chloroplast SRP/FtsY and Alb3 in protein integration into the thylakoid membrane. in the enzymes, vol 25, 3rd edn. In: Dalbey RE, Koehler CM Tamanoi F (eds) Molecular machines involved in protein transport across cellular membranes. Academic Press, San Diego, pp 493–521
Serek J, Bauer-Manz G, Struhalla G, Van Den Berg L, Kiefer D, Dalbey R, Kuhn A (2004) Escherichia coli YidC is a membrane insertase for Sec-independent proteins. EMBO J 23:294–301
Scotti PA, Urbanus ML, Brunner J, de Gier JW, von Heijne G, van der Does C, Driessen AJ, Oudega B, Luirink J (2000) YidC, the Escherichia coli homologue of mitochondrial Oxa1p, is a component of the Sec translocase. EMBO J 19:542–549
Samuelson JC, Jiang F, Yi L, Chen M, de Gier JW, Kuhn A, Dalbey RE (2001) Function of YidC for the insertion of M13 procoat protein in E. coli: translocation of mutants that show differences in their membrane potential dependence and Sec- requirement. J Biol Chem 16:16
Nouwen N, Driessen AJ (2002) SecDFyajC forms a heterotetrameric complex with YidC. Mol Microbiol 44:1397–1405
Xie K, Kiefer D, Nagler G, Dalbey RE, Kuhn A (2006) Different regions of the nonconserved large periplasmic domain of Escherichia coli YidC are involved in the SecF interaction and membrane insertase activity. Biochemistry 45:13401–13408
Xie K, Dalbey RE (2008) Inserting proteins into the bacterial cytoplasmic membrane using the Sec and YidC translocases. Nat Rev Microbiol 6:234–244
Kol S, Nouwen N, Driessen AJ (2008) Mechanisms of YidC-mediated insertion and assembly of multimeric membrane protein complexes. J Biol Chem 3505–3525
van der Laan M, Nouwen NP, Driessen AJ (2005) YidC–an evolutionary conserved device for the assembly of energy-transducing membrane protein complexes. Curr Opin Microbiol 8:182–187
Saaf A, Monne M, de Gier JW, von Heijne G (1998) Membrane topology of the 60-kDa Oxa1p homologue from Escherichia coli. J Biol Chem 273:30415–30418
Oliver DC, Paetzel M (2008) Crystal structure of the major periplasmic domain of the bacterial membrane protein assembly facilitator YidC. J Biol Chem 283:5208–5216
Ravaud S, Stjepanovic G, Wild K, Sinning I (2008) The crystal structure of the periplasmic domain of the Escherichia coli membrane protein insertase YidC contains a substrate binding cleft. J Biol Chem 283:9350–9358
Yu Z, Koningstein G, Pop A, Luirink J (2008) The conserved third transmembrane segment of YidC contacts nascent Escherichia coli inner membrane proteins. J Biol Chem 283:34635–34642
Klenner C, Yuan J, Dalbey RE, Kuhn A (2008) The Pf3 coat protein contacts TM1 and TM3 of YidC during membrane biogenesis. FEBS Lett 582:3967–3972
Winterfeld S, Imhof N, Roos T, Bar G, Kuhn A, Gerken U (2009) Substrate-induced conformational change of the Escherichia coli membrane insertase YidC. Biochemistry 48:6684–6691
van der Laan M, Houben EN, Nouwen N, Luirink J, Driessen AJ (2001) Reconstitution of Sec-dependent membrane protein insertion: nascent FtsQ interacts with YidC in a SecYEG-dependent manner. EMBO Rep 2:519–523
Nargang FE, Preuss M, Neupert W, Herrmann JM (2002) The Oxa1 protein forms a homooligomeric complex and is an essential part of the mitochondrial export translocase in Neurospora crassa. J Biol Chem 277:12846–12853
Kohler R, Boehringer D, Greber B, Bingel-Erlenmeyer R, Collinson I, Schaffitzel C, Ban N (2009) YidC and Oxa1 form dimeric insertion pores on the translating ribosome. Mol Cell 34:344–353
Jiang F, Chen M, Yi L, de Gier JW, Kuhn A, Dalbey RE (2003) Defining the regions of Escherichia coli YidC that contribute to activity. J Biol Chem 278:48965–48972
Yuan J, Phillips GJ, Dalbey RE (2007) Isolation of cold-sensitive yidC mutants provides insights into the substrate profile of the YidC insertase and the importance of transmembrane 3 in YidC function. J Bacteriol 189:8961–8972
Yen MR, Harley KT, Tseng YH, Saier MH Jr (2001) Phylogenetic and structural analyses of the oxa1 family of protein translocases. FEMS Microbiol Lett 204:223–231
Tjalsma H, Bron S, van Dijl JM (2003) Complementary impact of paralogous Oxa1-like proteins of Bacillus subtilis on post-translocational stages in protein secretion. J Biol Chem 278:15622–15632
Murakami T, Haga K, Takeuchi M, Sato T (2002) Analysis of the Bacillus subtilis spoIIIJ gene and its paralogue gene, yqjG. J Bacteriol 184:1998–2004
Camp AH, Losick R (2008) A novel pathway of intercellular signalling in Bacillus subtilis involves a protein with similarity to a component of type III secretion channels. Mol Microbiol 69:402–417
Serrano M, Vieira F, Moran CP Jr, Henriques AO (2008) Processing of a membrane protein required for cell-to-cell signaling during endospore formation in Bacillus subtilis. J Bacteriol 190:7786–7796
Dong Y, Palmer SR, Hasona A, Nagamori S, Kaback HR, Dalbey RE, Brady LJ (2008) Functional overlap but lack of complete cross-complementation of Streptococcus mutans and Escherichia coli YidC orthologs. J Bacteriol 190:2458–2469
Funes S, Hasona A, Bauerschmitt H, Grubbauer C, Kauff F, Collins R, Crowley PJ, Palmer SR, Brady LJ, Herrmann JM (2009) Independent gene duplications of the YidC/Oxa/Alb3 family enabled a specialized cotranslational function. Proc Natl Acad Sci USA 106:6656–6661
Natale P, Bruser T, Driessen AJ (2008) Sec- and Tat-mediated protein secretion across the bacterial cytoplasmic membrane-distinct translocases and mechanisms. Biochim Biophys Acta 1778:1735–1756
Palmer T, Sargent F, Berks BC (2005) Export of complex cofactor-containing proteins by the bacterial Tat pathway. Trends Microbiol 13:175–180
Robinson C, Bolhuis A (2004) Tat-dependent protein targeting in prokaryotes and chloroplasts. Biochim Biophys Acta 1694:135–147
Sargent F (2007) The twin-arginine transport system: moving folded proteins across membranes. Biochem Soc Trans 35:835–847
Hatzixanthis K, Palmer T, Sargent F (2003) A subset of bacterial inner membrane proteins integrated by the twin-arginine translocase. Mol Microbiol 49:1377–1390
Dilks K, Gimenez MI, Pohlschroder M (2005) Genetic and biochemical analysis of the twin-arginine translocation pathway in halophilic archaea. J Bacteriol 187:8104–8113
Widdick DA, Dilks K, Chandra G, Bottrill A, Naldrett M, Pohlschroder M, Palmer T (2006) The twin-arginine translocation pathway is a major route of protein export in Streptomyces coelicolor. Proc Natl Acad Sci USA 103:17927–17932
Widdick DA, Eijlander RT, van Dijl JM, Kuipers OP, Palmer T (2008) A facile reporter system for the experimental identification of twin-arginine translocation (Tat) signal peptides from all kingdoms of life. J Mol Biol 375:595–603
Saint-Joanis B, Demangel C, Jackson M, Brodin P, Marsollier L, Boshoff H, Cole ST (2006) Inactivation of Rv2525c, a substrate of the twin arginine translocation (Tat) system of Mycobacterium tuberculosis, increases beta-lactam susceptibility and virulence. J Bacteriol 188:6669–6679
Posey JE, Shinnick TM, Quinn FD (2006) Characterization of the twin-arginine translocase secretion system of Mycobacterium smegmatis. J Bacteriol 188:1332–1340
McDonough JA, Hacker KE, Flores AR, Pavelka MS Jr, Braunstein M (2005) The twin-arginine translocation pathway of Mycobacterium smegmatis is functional and required for the export of mycobacterial beta-lactamases. J Bacteriol 187:7667–7679
Bronstein PA, Marrichi M, Cartinhour S, Schneider DJ, DeLisa MP (2005) Identification of a twin-arginine translocation system in Pseudomonas syringae pv. tomato DC3000 and its contribution to pathogenicity and fitness. J Bacteriol 187:8450–8461
Caldelari I, Mann S, Crooks C, Palmer T (2006) The Tat pathway of the plant pathogen Pseudomonas syringae is required for optimal virulence. Mol Plant Microbe Interact 19:200–212
Lavander M, Ericsson SK, Broms JE, Forsberg A (2006) The twin arginine translocation system is essential for virulence of Yersinia pseudotuberculosis. Infect Immun 74:1768–1776
De Buck E, Maes L, Meyen E, Van Mellaert L, Geukens N, Anne J, Lammertyn E (2005) Legionella pneumophila Philadelphia-1 tatB and tatC affect intracellular replication and biofilm formation. Biochem Biophys Res Commun 331:1413–1420
Rossier O, Cianciotto NP (2005) The Legionella pneumophila tatB gene facilitates secretion of phospholipase C, growth under iron-limiting conditions, and intracellular infection. Infect Immun 73:2020–2032
De Buck E, Lammertyn E, Anne J (2008) The importance of the twin-arginine translocation pathway for bacterial virulence. Trends Microbiol 16:442–453
Cristobal S, de Gier JW, Nielsen H, von Heijne G (1999) Competition between Sec- and TAT-dependent protein translocation in Escherichia coli. EMBO J 18:2982–2990
Tullman-Ercek D, DeLisa MP, Kawarasaki Y, Iranpour P, Ribnicky B, Palmer T, Georgiou G (2007) Export pathway selectivity of Escherichia coli twin arginine translocation signal peptides. J Biol Chem 282:8309–8316
Blaudeck N, Kreutzenbeck P, Freudl R, Sprenger GA (2003) Genetic analysis of pathway specificity during posttranslational protein translocation across the Escherichia coli plasma membrane. J Bacteriol 185:2811–2819
Bogsch E, Brink S, Robinson C (1997) Pathway specificity for a delta pH-dependent precursor thylakoid lumen protein is governed by a ‘Sec-avoidance’ motif in the transfer peptide and a ‘Sec-incompatible’ mature protein. EMBO J 16:3851–3859
Cristobal S, Scotti P, Luirink J, von Heijne G, de Gier JW (1999) The signal recognition particle-targeting pathway does not necessarily deliver proteins to the sec-translocase in Escherichia coli. J Biol Chem 274:20068–20070
Ize B, Gerard F, Wu LF (2002) In vivo assessment of the Tat signal peptide specificity in Escherichia coli. Arch Microbiol 178:548–553
Wu LF, Ize B, Chanal A, Quentin Y, Fichant G (2000) Bacterial twin-arginine signal peptide-dependent protein translocation pathway: evolution and mechanism. J Mol Microbiol Biotechnol 2:179–189
Behrendt J, Standar K, Lindenstrauss U, Bruser T (2004) Topological studies on the twin-arginine translocase component TatC. FEMS Microbiol Lett 234:303–308
Ki JJ, Kawarasaki Y, Gam J, Harvey BR, Iverson BL, Georgiou G (2004) A periplasmic fluorescent reporter protein and its application in high-throughput membrane protein topology analysis. J Mol Biol 341:901–909
Sargent F, Gohlke U, De Leeuw E, Stanley NR, Palmer T, Saibil HR, Berks BC (2001) Purified components of the Escherichia coli Tat protein transport system form a double-layered ring structure. Eur J Biochem 268:3361–3367
Gohlke U, Pullan L, McDevitt CA, Porcelli I, de Leeuw E, Palmer T, Saibil HR, Berks BC (2005) The TatA component of the twin-arginine protein transport system forms channel complexes of variable diameter. Proc Natl Acad Sci USA 102:10482–10486
Oates J, Barrett CM, Barnett JP, Byrne KG, Bolhuis A, Robinson C (2005) The Escherichia coli twin-arginine translocation apparatus incorporates a distinct form of TatABC complex, spectrum of modular TatA complexes and minor TatAB complex. J Mol Biol 346:295–305
Alami M, Luke I, Deitermann S, Eisner G, Koch HG, Brunner J, Muller M (2003) Differential interactions between a twin-arginine signal peptide and its translocase in Escherichia coli. Mol Cell 12:937–946
Bolhuis A, Mathers JE, Thomas JD, Barrett CM, Robinson C (2001) TatB and TatC form a functional and structural unit of the twin-arginine translocase from Escherichia coli. J Biol Chem 276:20213–20219
Holzapfel E, Eisner G, Alami M, Barrett CM, Buchanan G, Luke I, Betton JM, Robinson C, Palmer T, Moser M, Muller M (2007) The entire N-terminal half of TatC is involved in twin-arginine precursor binding. Biochemistry 46:2892–2898
Blaudeck N, Kreutzenbeck P, Muller M, Sprenger GA, Freudl R (2005) Isolation and characterization of bifunctional Escherichia coli TatA mutant proteins that allow efficient tat-dependent protein translocation in the absence of TatB. J Biol Chem 280:3426–3432
Barnett JP, Eijlander RT, Kuipers OP, Robinson C (2008) A minimal Tat system from a gram-positive organism: a bifunctional TatA subunit participates in discrete TatAC and TatA complexes. J Biol Chem 283:2534–2542
Jongbloed JD, van der Ploeg R, van Dijl JM (2006) Bifunctional TatA subunits in minimal Tat protein translocases. Trends Microbiol 14:2–4
Jongbloed JD, Grieger U, Antelmann H, Hecker M, Nijland R, Bron S, van Dijl JM (2004) Two minimal Tat translocases in Bacillus. Mol Microbiol 54:1319–1325
Jongbloed JD, Antelmann H, Hecker M, Nijland R, Bron S, Airaksinen U, Pries F, Quax WJ, van Dijl JM, Braun PG (2002) Selective contribution of the twin-arginine translocation pathway to protein secretion in Bacillus subtilis. J Biol Chem 277:44068–44078
Kouwen TR, van der Ploeg R, Antelmann H, Hecker M, Homuth G, Mader U, van Dijl JM (2009) Overflow of a hyper-produced secretory protein from the Bacillus Sec pathway into the Tat pathway for protein secretion as revealed by proteogenomics. Proteomics 9:1018–1032
Sargent F, Stanley NR, Berks BC, Palmer T (1999) Sec-independent protein translocation in Escherichia coli: a distinct and pivotal role for the TatB protein. J Biol Chem 274:36073–36082
Bolhuis A (2002) Protein transport in the halophilic archaeon Halobacterium sp. NRC-1: a major role for the twin-arginine translocation pathway? Microbiology 148:3335–3346
Thomas JR, Bolhuis A (2006) The tatC gene cluster is essential for viability in halophilic archaea. FEMS Microbiol Lett 256:44–49
Gimenez MI, Dilks K, Pohlschroder M (2007) Haloferax volcanii twin-arginine translocation substates include secreted soluble, C-terminally anchored and lipoproteins. Mol Microbiol 66:1597–1606
Mould RM, Robinson C (1991) A proton gradient is required for the transport of two lumenal oxygen-evolving proteins across the thylakoid membrane. J Biol Chem 266:12189–12193
Braun NA, Davis AW, Theg SM (2007) The chloroplast Tat pathway utilizes the transmembrane electric potential as an energy source. Biophys J 93:1993–1998
Kwan DC, Thomas JR, Bolhuis A (2008) Bioenergetic requirements of a Tat-dependent substrate in the halophilic archaeon Haloarcula hispanica. FEBS J 275:6159–6167
Bageshwar UK, Musser SM (2007) Two electrical potential-dependent steps are required for transport by the Escherichia coli Tat machinery. J Cell Biol 179:87–99
Acknowledgments
This work was supported by the National Institutes of Health grant GM63862 (to R.E. D.), EU grants LSHM-CT-2006-019064 and PITN-GA-2008-215524, the transnational SysMO initiative through project BACELL SysMO, the European Science Foundation under the EUROCORES Programme EuroSCOPE, and grant 04-EScope 01-011 from the Research Council for Earth and Life Sciences of the Netherlands Organization for Scientific Research (to J.M.v.D.).
Author information
Authors and Affiliations
Corresponding authors
Additional information
J. Yuan and J. C. Zweers contributed equally to this work.
Rights and permissions
About this article
Cite this article
Yuan, J., Zweers, J.C., van Dijl, J.M. et al. Protein transport across and into cell membranes in bacteria and archaea. Cell. Mol. Life Sci. 67, 179–199 (2010). https://doi.org/10.1007/s00018-009-0160-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-009-0160-x