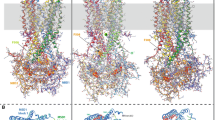
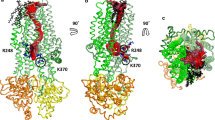
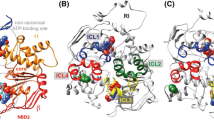
Abstract
Cystic fibrosis transmembrane conductance regulator (CFTR), involved in cystic fibrosis (CF), is a chloride channel belonging to the ATP-binding cassette (ABC) superfamily. Using the experimental structure of Sav1866 as template, we previously modeled the human CFTR structure, including membrane-spanning domains (MSD) and nucleotide-binding domains (NBD), in an outward-facing conformation (open channel state). Here, we constructed a model of the CFTR inward-facing conformation (closed channel) on the basis of the recent corrected structures of MsbA and compared the structural features of those two states of the channel. Interestingly, the MSD:NBD coupling interfaces including F508 (ΔF508 being the most common CF mutation) are mainly left unchanged. This prediction, completed by the modeling of the regulatory R domain, is supported by experimental data and provides a molecular basis for a better understanding of the functioning of CFTR, especially of the structural features that make CFTR the unique channel among the ABC transporters.








Similar content being viewed by others
References
Gadsby DC, Vergani P, Csanády L (2006) The ABC protein turned chloride channel whose failure causes cystic fibrosis. Nature 440:477–483
Rowe SM, Miller S, Sorscher EJ (2005) Cystic fibrosis. N Engl J Med 352:1992–2001
Biemans-Oldehinkel E, Doeven MK, Poolman B (2006) ABC transporter architecture and regulatory roles of accessory domains. FEBS Lett 580:1023–1035
Anderson MP, Gregory RJ, Thompson S, Souza DW, Paul S, Mulligan RC, Smith AE, Welsh MJ (1991) Demonstration that CFTR is a chloride channel by alteration of its anion selectivity. Science 253:202–205
Bear CE, Li CH, Kartner N, Bridges RJ, Jensen TJ, Ramjeesingh M, Riordan JR (1992) Purification and functional reconstitution of the cystic fibrosis transmembrane conductance regulator (CFTR). Cell 68:809–818
Gadsby DC (2009) Ion channels versus ion pumps: the principal difference, in principle. Nat Rev Mol Cell Biol 10:344–352
Cheng SH, Rich DP, Marshall J, Gregory RJ, Welsh MJ, Smith AE (1991) Phosphorylation of the R domain by cAMP-dependent protein kinase regulates the CFTR chloride channel. Cell 66:1027–1036
Ostedgaard LS, Baldursson O, Welsh MJ (2001) Regulation of the cystic fibrosis transmembrane conductance regulator Cl− channel by its R domain. J Biol Chem 276:7689–7692
Cheng SH, Gregory RJ, Marshall J, Paul S, Souza DW, White GA, O’Riordan CR, Smith AE (1990) Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell 63:827–834
Riordan JR (1999) Cystic fibrosis as a disease of misprocessing of the cystic fibrosis transmembrane conductance regulator glycoprotein. Am J Hum Genet 64:1499–1504
Ward CL, Omura S, Kopito RR (1995) Degradation of CFTR by the ubiquitin–proteasome pathway. Cell 83:121–127
Callebaut I, Eudes R, Mornon JP, Lehn P (2004) Nucleotide-binding domains of human cystic fibrosis transmembrane conductance regulator: detailed sequence analysis and three-dimensional modeling of the heterodimer. Cell Mol Life Sci 61:230–242
Eudes R, Lehn P, Férec C, Mornon J-P, Callebaut I (2005) Nucleotide binding domains of human CFTR: a structural classification of critical residues and disease-causing mutations. Cell Mol Life Sci 62:2112–2123
Lewis HA, Buchanan SG, Burley SK, Conners K, Dickey M, Dorwart M, Fowler R, Gao X, Guggino WB, Hendrickson WA, Hunt J, Kearins MC, Lorimer D, Maloney PC, Post KW, Rajashankar KR, Rutter ME, Sauder JM, Shriver S, Thibodeau PH, Thomas PJ, Zhang M, Zhao X, Emtage S (2004) Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator. EMBO J 23:282–293
Lewis HA, Zhao X, Wang C, Sauder JM, Rooney I, Noland BW, Lorimer D, Kearins MC, Conners K, Condon B, Maloney PC, Guggino WB, Hunt JF, Emtage S (2005) Impact of the deltaF508 mutation in first nucleotide-binding domain of human cystic fibrosis transmembrane conductance regulator on domain folding and structure. J Biol Chem 280:1346–1353
Cui L, Aleksandrov L, Chang XB, Hou YX, He L, Hegedus T, Gentzsch M, Aleksandrov A, Balch WE, Riordan JR (2007) Domain interdependence in the biosynthetic assembly of CFTR. J Mol Biol 365:981–994
Du K, Sharma M, Lukacs GL (2005) The DeltaF508 cystic fibrosis mutation impairs domain–domain interactions and arrests post-translational folding of CFTR. Nat Struct Mol Biol 12:17–25
Dawson RJ, Locher KP (2007) Structure of the multidrug ABC transporter Sav1866 from Staphylococcus aureus in complex with AMP–PNP. FEBS Lett 581:935–938
Mornon J-P, Lehn P, Callebaut I (2008) Atomic model of human cystic fibrosis transmembrane conductance regulator: membrane-spanning domains and coupling interfaces. Cell Mol Life Sci 65:2594–2612
Serohijos AW, Hegedus T, Aleksandrov AA, He L, Cui L, Dokholyan NV, Riordan JR (2008) Phenylalanine-508 mediates a cytoplasmic-membrane domain contact in the CFTR 3D structure crucial to assembly and channel function. Proc Natl Acad Sci USA 105:3256–3261
Loo TW, Bartlett MC, Clarke DM (2008) Processing mutations disrupt interactions between the nucleotide binding and transmembrane domains of P-glycoprotein and the cystic fibrosis transmembrane conductance regulator (CFTR). J Biol Chem 283:28190–28197
Aller SG, Yu J, Ward A, Weng Y, Chittaboina S, Zhuo R, Harrell PM, Trinh YT, Zhang Q, Urbatsch IL, Chang G (2009) Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science 323:1718–1722
Krasnov KV, Tzetis M, Cheng J, Guggino WB, Cutting GR (2008) Localization studies of rare missense mutations in cystic fibrosis transmembrane conductance regulator (CFTR) facilitate interpretation of genotype-phenotype relationships. Hum Mut 29:1364–1372
Seibert FS, Linsdell P, Loo TW, Hanrahan JW, Clarke DM, Riordan JR (1996) Disease-associated mutations in the fourth cytoplasmic loop of cystic fibrosis transmembrane conductance regulator compromise biosynthetic processing and chloride channel activity. J Biol Chem 271:15139–15145
Ward A, Reyes CL, Yu J, Roth CB, Chang G (2007) Flexibility in the ABC transporter MsbA: alternating access with a twist. Proc Natl Aca Sci USA 104:19005–19010
Oldham ML, Davidson AL, Chen J (2008) Structural insights into ABC transporter mechanism. Curr Opin Struct Biol 18:726–733
Rees DC, Johnson E, Lewinson O (2009) ABC transporters: the power to change. Nat Rev Mol Cell Biol 10:218–227
Mio K, Ogura T, Mio M, Shimizu H, Hwang T, Sato C, Sohma Y (2008) Three-dimensional reconstruction of human cystic fibrosis transmembrane conductance regulator chloride channel revealed an ellipsoidal structure with orifices beneath the putative transmembrane domain. J Biol Chem 283:30300–30310
Zhang L, Aleksandrov LA, Zhao Z, Birtley JR, Riordan JR, Ford RC (2009) Architecture of the cystic fibrosis transmembrane conductance regulator protein and structural changes associated with phosphorylation and nucleotide binding. J Struct Biol (in press)
Rosenberg MF, Kamis AB, Aleksandrov LA, Ford RC, Riordan JR (2004) Purification and crystallization of the cystic fibrosis transmembrane conductance regulator (CFTR). J Biol Chem 279:39051–39057
Marti-Renom MA, Stuart A, Fiser A, Sánchez R, Melo F, Sali A (2000) Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct 29:291–325
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723
Callebaut I, Labesse G, Durand P, Poupon A, Canard L, Chomilier J, Henrissat B, Mornon JP (1997) Deciphering protein sequence information through hydrophobic cluster analysis (HCA): current status and perspectives. Cell Mol Life Sci 53:621–645
Eudes R, Le Tuan K, Delettré J, Mornon JP, Callebaut I (2007) A generalized analysis of hydrophobic and loop clusters within globular protein sequences. BMC Struct Biol 7:2
Gaboriaud C, Bissery V, Benchetrit T, Mornon JP (1987) Hydrophobic cluster analysis: an efficient new way to compare and analyse amino acid sequences. FEBS Lett 224:149–155
Chen J, Lu G, Lin J, Davidson AL, Quiocho FA (2003) A tweezers-like motion of the ATP-binding cassette dimer in an ABC transport cycle. Mol Cell 12:651–661
Fatehi M, Linsdell P (2009) Novel residues lining the CFTR chloride channel pore identified by functional modification of introduced cysteines. J Membr Biol 228:151–164
Guinamard R, Akabas MH (1999) Arg352 is a major determinant of charge selectivity in the cystic fibrosis transmembrane conductance regulator chloride channel. Biochemistry 38:5528–5537
Linsdell P, Zheng SX, Hanrahan JW (1998) Non-pore lining amino acid side chains influence anion selectivity of the human CFTR Cl− channel expressed in mammalian cell lines. J Physiol 512:1–16
McDonough S, Davidson N, Lester HA, McCarty NA (1994) Novel pore-lining residues in CFTR that govern permeation and open-channel block. Neuron 13:623–634
Gong X, Linsdell P (2003) Molecular determinants and role of an anion binding site in the external mouth of the CFTR chloride channel pore. J Physiol 549:387–397
Zhou JJ, Fatehi M, Linsdell P (2008) Identification of positive charges situated at the outer mouth of the CFTR chloride channel pore. Pflugers Arch 457:351–360
Ge N, Muise CN, Gong X, Linsdell P (2004) Direct comparison of the functional roles played by different transmembrane regions in the cystic fibrosis transmembrane conductance regulator chloride channel pore. J Biol Chem 279:55283–55289
Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou JL et al (1989) Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science 245:1066–1073
St Aubin CN, Linsdell P (2006) Positive charges at the intracellular mouth of the pore regulate anion conduction in the CFTR chloride channel. J Gen Physiol 128:535–545
St Aubin CN, Zhou JJ, Linsdell P (2007) Identification of a second blocker binding site at the cytoplasmic mouth of the cystic fibrosis transmembrane conductance regulator chloride channel pore. Mol Pharmacol 71:1360–1368
Baker JM, Hudson RP, Kanelis V, Choy WY, Thibodeau PH, Thomas PJ, Forman-Kay JD (2007) CFTR regulatory region interacts with NBD1 predominantly via multiple transient helices. Nat Struct Mol Biol 14:738–745
Hegedus T, Serohijos AW, Dokholyan NV, He L, Riordan JR (2008) Computational studies reveal phosphorylation-dependent changes in the unstructured R domain of CFTR. J Mol Biol 378:1052–1063
Gros P, Croop J, Housman D (1986) Mammalian multidrug resistance gene: complete cDNA sequence indicates strong homology to bacterial transport proteins. Cell 47:371–380
Diederichs K, Diez J, Greller G, Müller C, Breed J, Schnell C, Vonrhein C, Boos W, Welte W (2000) Crystal structure of MalK, the ATPase subunit of the trehalose/maltose ABC transporter of the archaeon Thermococcus litoralis. EMBO J 19:5951–5961
Hollenstein K, Dawson JP, Locker KP (2007) Structure and mechanism of ABC transporter proteins. Curr Opin Struct Biol 17:412–418
Koonin EV, Wolf Y, Aravind L (2000) Protein fold recognition using sequence profiles and its application in structural genomics. Adv Protein Chem 54:245–275
Verdon G, Albers SV, Dijkstra BW, Driessen AJ, Thunnissen AM (2003) Crystal structures of the ATPase subunit of the glucose ABC transporter from Sulfolobus solfataricus: nucleotide-free and nucleotide-bound conformations. J Mol Biol 330:343–358
Awayn NH, Rosenberg MF, Kamis AB, Aleksandrov LA, Riordan JR, Ford RC (2005) Crystallographic and single-particle analyses of native- and nucleotide-bound forms of the cystic fibrosis transmembrane conductance regulator (CFTR) protein. Biochem Soc Trans 33:996–999
Zhou Z, Wang X, Liu HY, Zou X, Li M, Hwang TC (2006) The two ATP binding sites of cystic fibrosis transmembrane conductance regulator (CFTR) play distinct roles in gating kinetics and energetics. J Gen Physiol 128:413–422
Hegedűs T, Aleksandrov A, Mengos A, Cui L, Jensen TJ, Riordan JR (2009) Role of individual R domain phosphorylation sites in CFTR regulation by protein kinase A. Biochim Biophys Acta 1788:1341–1349
Rich DP, Gregory RJ, Anderson MP, Manavalan P, Smith AE, Welsh MJ (1991) Effect of deleting the R domain on CFTR-generated chloride channels. Science 253:205–207
Chappe V, Irvine T, Liao J, Evagelidis A, Hanrahan JW (2005) Phosphorylation of CFTR by PKA promotes binding of the regulatory domain. EMBO J 24:2730–2740
Kos V, Ford RC (2009) The ATP-binding cassette family: a structural perspective. Cell Mol Life Sci (in press)
Nishimura K, Kim S, Zhang L, Cross T (2002) The closed state of a H+ channel helical bundle combining precise orientational and distance restraints from solid state NMR. Biochemistry 41:13170–13177
Dutzler R, Campbell EB, MacKinnon R (2003) Gating the selectivity filter in ClC chloride channels. Science 300:108–112
Doyle DA (2004) Structural changes during ion channel gating. Trends Neurosci 27:298–302
Fu J, Kirk KL (2001) Cysteine substitutions reveal dual functions of the amino-terminal tail in cystic fibrosis transmembrane conductance regulator channel gating. J Biol Chem 276:35660–35668
Naren AP, Cormet-Boyaka E, Fu J, Villain M, Blalock JE, Quick MW, Kirk KL (1999) CFTR chloride channel regulation by an interdomain interaction. Science 286:544–548
Gouet P, Courcelle E, Stuart DI, Metoz F (1999) ESPript: multiple sequence alignments in PostScript. Bioinformatics 15:305–308
Lobet S, Dutzler R (2006) Ion-binding properties of the ClC chloride selectivity filter. EMBO J 25:24–33
Acknowledgments
This work was supported by grants from the association "Vaincre La Mucoviscidose" (Paris, France).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1: Modeling of the inward-facing conformation of CFTR (closed channel).
The passage from the MSD configuration found in the MsbA closed apo model to a “tight” closed apo conformation, likely representing the closed form of the CFTR channel, is illustrated here. This occurs through a small rigid body rotation of the two MSDs around the ICLs pivots. (TIFF 999 kb)
Figure S2: Superimposition of the MsbA open apo conformer with the tight conformation of the CFTR inward-facing model, in the vicinity of the channel entrance (extracellular side) and showing the transmembrane helices
. The transmembrane helices of the MsbA open apo conformer are depicted in grey and are labeled TM1 to TM12, whereas those of the tight conformer (colored) are labeled TM1t to TM12t. One can note the good overall correspondence between the two conformers, noticeably for helices TM6 and TM12 at the center. (TIFF 433 kb)
Figure S3: Detailed views of the CFTR chloride channel in the “tight”, inward-facing conformation. A) Side view (left) and top view (right); B) Stereo views from the top.
The orientations are similar to those shown in Figure 3. The potential chloride translocation pathway is symbolized by colored spheres (the colors ranging from blue to red while moving from the intracellular side to the extracellular side). (TIFF 740 kb)
Figure S4:
The R2 domain: Sequence alignment deduced from the HCA comparison of the C-terminal part of the CFTR R domain (here named the R2 domain), preceding the second membrane-spanning domain (starting at transmembrane helix TM7), with the N-terminal tail of the same protein, preceding the first membrane-spanning domain (MSD1, starting at transmembrane helix TM1). The CFTR sequences of four species are shown: Homo sapiens (h, genbank identifier (gi): 147744553), Salmo salar (s, gi:12746235), Danio rerio (d, gi:117380075), Fundulus heteroclitus (f, gi:3015540), illustrating the presently known most divergent CFTR sequences. At top are presented a few N-terminal sequences of other ABC proteins, sharing significant similarities with the CFTR N-terminal sequence (hABCCB: Homo sapiens ABCC11, gi:74762666; hMRP9: Homo sapiens MRP9, gi:161788999; hMRP5: Homo sapiens MRP5, gi:8928547; dGD10651: Drosophila simulans GD10651, gi:195581868, hMRP4: Homo sapiens MRP4, gi:206729914). Between the CFTR N-terminal and R2 sequences are shown the N-terminal sequence of Saccharomyces cerevisiae YOR1 (yYOR1, gi:1730876) and the R sequence of Homo sapiens MRP3 (hMRP3: gi:6920069), which provide intermediate links. The alanine content of the region aligned with the N-ter helix is in good agreement with such a secondary structure. Conserved hydrophobic amino acids (V, I, L, F, M, Y, W) are colored dark green, and residues that can substitute them (A, C, T) are colored light green. Loop-forming amino acids (P, G, D, N, S) are colored yellow. Conserved aromatic residues (Y, W, F) are colored purple, conserved basic and acidic residues are colored blue and pink, respectively. The secondary structures predicted using HCA for the associated hydrophobic clusters (1 stands for V, I, L, F, M, Y, W; 0 for other residues (Eudes et al., BMC Struct Biol (2007) 7: 2) are indicated above and below the CFTR N-terminal and R2 sequence, respectively. Black stars indicate the phosphorylation sites. The amino acid ranges are shown at the end of the sequences. Below the alignment are shown the secondary structure propensities estimated for the R2 domain, based on NMR experimental data, for the non-phosphorylated and phosphorylated forms of CFTR (based on the data presented in Figure 2 of the Baker’s article (Baker et al., Nat Struct Mol Biol (2007) 14: 738-745). (TIFF 517 kb)
Figure S5: 3D structures of the regulatory domains (TOBE domains) of three ABC proteins.
From left to right: the Pyrococcus horikoshii multiple sugar binding transport ATP-binding protein (pdb 2d62), the Archaeoglobus fulgidus molybdate/tungstate ABC transporter ModC (pdb 3d31) and the Sulfolobus solfataricus glucose transporter (pdb 1oxs). Strands constituting the “Greek-Key” motifs are labeled according to Figure S6. The fifth strand of each TOBE domain is depicted in pink. The N- and C-termini of the regulatory domains are indicated. The WO4 groups, which bind to the ModC regulatory domains and allow a trans-inhibition of the molybdate/tungstate ABC transporter, are shown. (TIFF 356 kb)
Figure S6: The R1 domain:
Top) Sequence alignment deduced from the HCA comparison of the N-terminal part of the CFTR R domain (here named the R1 domain) with the regulatory domains found in some bacterial ABC transporters immediately after their NBDs (TOBE domains). The sequences of six of these proteins are shown, distributed into two subfamilies. The similarities between amino acids are reported as in Figure S4, and phosphorylation sites are indicated by black stars. Grey stars indicate the C-terminal amino acid. The position of the regulatory extension (RE), observed in the NBD1 crystal structure, is indicated. Bottom) Secondary structure propensities estimated for the R1 domain, based on NMR experimental data, for the non-phosphorylated and phosphorylated forms of CFTR (based on the data presented in Figure 2 of the Baker’s article (Baker et al., Nat Struct Mol Biol (2007) 14: 738-745). (TIFF 407 kb)
Figure S7: Comparison of the HCA plots of the human CFTR R domain (aa 610 to 885) and of the yeast Yor1p corresponding region (aa 775 to 925).
The sequence is shown on a duplicated alpha-helical net, and the strong hydrophobic amino acids (V I L F M Y W) are contoured: these form hydrophobic clusters, which mainly correspond to regular secondary structures. Guidelines to the use of the method can be found in Gaboriaud et al.FEBS Lett (1987) 224: 149-155 and in Callebaut et al. (1997) Cell Mol Life Sci53: 621-645. The predicted secondary structures and domain limits are reported above the human CFTR plot. Green colors highlights conservation of hydrophobic amino acids (V I L F M Y W), whereas orange is used for correspondence between hydrophobic amino acids (V I L F M Y W) and amino acids (A C T), which can substitute them in a context-dependent manner. Sequence identities are colored pink. (TIFF 197 kb)
Figure S9: Putative dimeric architecture of CFTR (closed, inward-facing conformation): an alternative solution to that proposed in Figure 5.
The view is shown in a similar orientation as in Figure 5. The interface between each monomer is built of the opposite and symmetric long faces, as in Figure 5 (compare the bottom views, where CFTR is seen from the extracellular side). Main contacts are between TM4 and TM4, αα2 of NBD2 and R2. However, in this alternative arrangement, R1 does not contact R1 (in the context of the proposed location of this domain). (TIFF 3510 kb)
Figure S10:
Possible concerted movements of the NBD1/NBD2 domains and the R1 and R2 parts of the regulatory domain R, between the inward-facing conformation (closed state) at left and the outward-facing conformation (open state, in which the bound nucleotides are shown). The NBD1 (blue) being fixed, the NBD2 domain rubs, while moving, the R2 and R1 segments and may interact more tightly with phosphorylable segments (S660, S737, S795 and S813 are labeled in red to recall their importance (Hegedüs et al. Biochim Biophys Acta (2009) 1788: 1341-1349)) in the channel open-state through specific basic patches. To that aim, R1 (in white) may rotate around its N- and C-terminal parts toward a position (in yellow) close to the NBD2 domain (in orange). See Fig. S11 for the details of amino acids in the R1 region for the outward-facing conformation. Red stars indicate the possible area of interaction. One can note that no phosphorylable serine is present after S813 within the R2 region which provides less contact with NBD2. (TIFF 1261 kb)
Figure S11:
Zoom on the model of the R1 domain (outward-facing conformation), in a position where it begins to interact with the two basic specific patches including K1429, R1434, R1438 (NBD2) and K1457, K1459, K1461 (C-ter domain). (TIFF 789 kb)
Figure S12:
Putative alternative pathway towards the entry of the CFTR channel pore through the NBD1:NBD2 interface, leading to the bundle of intracellular loops (outward-facing configuration (Mornon et al. (2008) Cell Mol Life Sci65: 2594-2612). A) Global view with, at the center in the middle distance, the cytoplasmic ends of ICL1 and ICL2 shown in dark blue and ICL3 and ICL4 shown in red. The R1 domain, which is likely situated in front of this view, is not shown. The R1 domain, with the position shown in Figure S10, however does not impede access to this pathway. B) Closer view, in the same direction as in panel A, but showing only the putative conduction pathway of the NBD1:NBD2 interface. Anions might be attracted by the basic residues K606 and R1403 and then transported through a small pore including G576 and A1374, which are 5.1 Å apart in the model, within the Walker B loops (βc4 -αc4-5 loop). They might leave the NBD1:NBD2 interface at the levels of the Q loops (Q493 and Q1291, which are 5.6 Å apart from each other). Basic residues are shown in blue, hydrophobic ones in green and hydrophilic ones in pink. Proline residues are shown in light yellow. A1374 and G576 are shown as van der Waals surfaces, with between them a chloride ion. C) Same view as in Figure 7B, but with an alternative position of E267 at the foreground, which may weakly interact with K968 in one of its possible location. (TIFF 1.33 mb)
Table 1: Comparison of the amino acid content of R “linkers” from some ABC proteins, in the increasing order of the percentage in strong hydrophobic amino acids (V, I, L, F, M, Y, W).
l indicates the length of the considered segment (in amino acids). The absolute numbers (nH) and percentages (%H) in hydrophobic amino acids (V, I, L, F, M, Y, W) are given in the following columns, as well as the absolute number (nL) of loop-forming amino acids (P, G, D, N, S) and the ratio between the number of hydrophobic and loop-forming amino acids (R = nH/nL). The CFTR R1 sub-domain, the CFTR R1-R2 and the CFTR R2 sub-domain have values between those encountered for typical disordered regions (e.g. the R region observed in the crystal structure of MDR/P-gp (Aller et al., Science (2009) 323: 1718-1722) and for canonical, folded globular domains (e.g. the NBD1 and NBD2 domains, which have 33 % of strong hydrophobic amino acids). R1 is here considered to start at aa 646 (after the helix α8 observed in the crystal structure of Glcv, see Figure S6) and R2 to end at amino acid 840, before the so-called N-helix, see Figure S4). The two last lines of the table refer to the R domains of ABCA1, the phosphorylation of which also regulating the protein function (Roosbeek et al., J Biol Chem (2004) 279: 37779-37788). (PPT 528 kb)
Rights and permissions
About this article
Cite this article
Mornon, JP., Lehn, P. & Callebaut, I. Molecular models of the open and closed states of the whole human CFTR protein. Cell. Mol. Life Sci. 66, 3469–3486 (2009). https://doi.org/10.1007/s00018-009-0133-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-009-0133-0