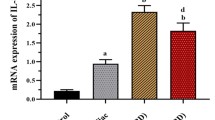
Abstract
Objective and design
Celiac disease (CD) is an intestinal inflammatory disorder of the small intestine. Gliadins are a component of gluten and there are three main types (α, γ, and ω). Recent studies indicate that gliadin peptides are able to activate an innate immune response. IL15 is a major mediator of the innate immune response and is involved in the early alteration of CD mucosa. The chitinase molecules are highly expressed by the innate immune cells during the inflammatory processes.
Material or subjects
We analyzed several microarray datasets of PBMCs and duodenum biopsies of CD patients and healthy control subjects (HCs). We verified the modulation CHI3L1 in CD patients and correlated the expression levels to the IL15, IL15Rα, TGM2, IFNγ, and IFNGR1/2. Duodenal biopsy samples belonged to nine active and nine treated children patients (long-term effects of gliadin), and 17 adult CD patients and 10 adults HCs. We also selected 169 samples of PBMCs from 127 CD patients on adherence to a gluten-free diet (GFD) for at least 2 years and 44 HCs.
Results
Our analysis showed that CHI3L1 and IL15Rα were significantly upregulated in adult and children’s celiac duodenum biopsies. In addition, the two genes were correlated significantly both in children than in adults CD duodenum biopsies. No significant modulation was observed in PBMCs of adult CD patients compared to the HCs. The correlation analysis of the expression levels of CHI3L1 and IL15Rα compared to TGM showed significant values both in adults and in children duodenal biopsies. Furthermore, the IFNγ expression levels were positively correlated with CHI3L1 and IL15Rα. Receiver operating characteristic (ROC) analysis confirmed the diagnostic ability of CHI3L1 and IL15Rα to discriminate CD from HCs.
Conclusion
Our data suggest a role for CHI3L1 underlying the pathophysiology of CD and represent a starting point aiming to inspire new investigation that proves the possible use of CHI3L1 as a diagnostic factor and therapeutic target.







Similar content being viewed by others
Abbreviations
- CD:
-
Celiac disease
- LP:
-
Lamina propria
- CHIT1:
-
Chitotriosidase
- CHIA:
-
Acidic mammalian chitinase
- CHI3L1:
-
Chitinase 3-like-1
- CHI3L2:
-
Chitinase 3-like-2
- CHID1:
-
Chitinase domain-containing 1
- IL15:
-
Interleukin 15
- (tTG)(TGM2):
-
Tissue transglutaminase
- APC:
-
Antigen presenting cells
- IFNγ:
-
Interferon gamma
References
Stenman SM, Lindfors K, Korponay-Szabo IR, Lohi O, Saavalainen P, Partanen J, et al. Secretion of celiac disease autoantibodies after in vitro gliadin challenge is dependent on small-bowel mucosal transglutaminase 2-specific IgA deposits. BMC Immunol. 2008;9:6.
Schuppan D, Junker Y, Barisani D. Celiac disease: from pathogenesis to novel therapies. Gastroenterology. 2009;137:1912–33.
Sollid LM, Jabri B. Triggers and drivers of autoimmunity: lessons from coeliac disease. Nat Rev Immunol. 2013;13:294–302.
Barbagallo I, Tibullo D, Di Rosa M, Giallongo C, Palumbo GA, Raciti G, et al. A cytoprotective role for the heme oxygenase-1/CO pathway during neural differentiation of human mesenchymal stem cells. J Neurosci Res. 2008;86:1927–35.
Stefanson AL, Bakovic M. Falcarinol is a potent inducer of heme oxygenase-1 and was more effective than sulforaphane in attenuating intestinal inflammation at diet-achievable doses. Oxid Med Cell Longev. 2018;2018:3153527.
Fernandez-Jimenez N, Castellanos-Rubio A, Plaza-Izurieta L, Irastorza I, Elcoroaristizabal X, Jauregi-Miguel A, et al. Coregulation and modulation of NFkappaB-related genes in celiac disease: uncovered aspects of gut mucosal inflammation. Hum Mol Genet. 2014;23:1298–310.
Maiuri MC, De Stefano D, Mele G, Fecarotta S, Greco L, Troncone R, et al. Nuclear factor kappa B is activated in small intestinal mucosa of celiac patients. J Mol Med (Berl). 2003;81:373–9.
Castillo EF, Schluns KS. Regulating the immune system via IL-15 transpresentation. Cytokine. 2012;59:479–90.
Furst DE, Emery P. Rheumatoid arthritis pathophysiology: update on emerging cytokine and cytokine-associated cell targets. Rheumatology. 2014;53:1560–9.
Rentzos M, Rombos A. The role of IL-15 in central nervous system disorders. Acta Neurol Scand. 2012;125:77–82.
Michalak-Stoma A, Pietrzak A, Szepietowski JC, Zalewska-Janowska A, Paszkowski T, Chodorowska G. Cytokine network in psoriasis revisited. Eur Cytokine Netw. 2011;22:160–8.
Jabri B, de Serre NP, Cellier C, Evans K, Gache C, Carvalho C, et al. Selective expansion of intraepithelial lymphocytes expressing the HLA-E-specific natural killer receptor CD94 in celiac disease. Gastroenterology. 2000;118:867–79.
Mention JJ, Ben Ahmed M, Begue B, Barbe U, Verkarre V, Asnafi V, et al. Interleukin 15: a key to disrupted intraepithelial lymphocyte homeostasis and lymphomagenesis in celiac disease. Gastroenterology. 2003;125:730–45.
Escudero-Hernandez C, Plaza-Izurieta L, Garrote JA, Bilbao JR, Cegec, Arranz E. Association of the IL-15 and IL-15Ralpha genes with celiac disease. Cytokine. 2017;99:73–9.
Maiuri L, Ciacci C, Auricchio S, Brown V, Quaratino S, Londei M. Interleukin 15 mediates epithelial changes in celiac disease. Gastroenterology. 2000;119:996–1006.
Di Sabatino A, Ciccocioppo R, Cupelli F, Cinque B, Millimaggi D, Clarkson MM, et al. Epithelium derived interleukin 15 regulates intraepithelial lymphocyte Th1 cytokine production, cytotoxicity, and survival in coeliac disease. Gut. 2006;55:469–77.
Chirdo FG, Millington OR, Beacock-Sharp H, Mowat AM. Immunomodulatory dendritic cells in intestinal lamina propria. Eur J Immunol. 2005;35:1831–40.
Macpherson AJ, Smith K. Mesenteric lymph nodes at the center of immune anatomy. J Exp Med. 2006;203:497–500.
DePaolo RW, Abadie V, Tang F, Fehlner-Peach H, Hall JA, Wang W, et al. Co-adjuvant effects of retinoic acid and IL-15 induce inflammatory immunity to dietary antigens. Nature. 2011;471:220–4.
Jabri B, Sollid LM. Tissue-mediated control of immunopathology in coeliac disease. Nat Rev Immunol. 2009;9:858–70.
Abadie V, Kim SM, Lejeune T, Palanski BA, Ernest JD, Tastet O, et al. IL-15, gluten and HLA-DQ8 drive tissue destruction in coeliac disease. Nature. 2020;578:600–4.
Herrera-Estrella A, Chet I. Chitinases in biological control. Exs. 1999;87:171–84.
Boot RG, Blommaart EF, Swart E, Ghauharali-van der Vlugt K, Bijl N, Moe C, et al. Identification of a novel acidic mammalian chitinase distinct from chitotriosidase. J Biol Chem. 2001;276:6770–8.
Di Rosa M, Malaguarnera G, De Gregorio C, D'Amico F, Mazzarino MC, Malaguarnera L. Modulation of chitotriosidase during macrophage differentiation. Cell Biochem Biophys. 2013;66:239–47.
Malaguarnera L, Ohazuruike LN, Tsianaka C, Antic T, Di Rosa M, Malaguarnera M. Human chitotriosidase polymorphism is associated with human longevity in Mediterranean nonagenarians and centenarians. J Hum Genet. 2010;55:8–12.
Di Rosa M, Tibullo D, Cambria D, Distefano G, Saccone S, Di Raimondo F, et al. Chitotriosidase expression during monocyte-derived dendritic cells differentiation and maturation. Inflammation. 2015;38:2082–91.
van Aalten DM, Komander D, Synstad B, Gaseidnes S, Peter MG, Eijsink VG. Structural insights into the catalytic mechanism of a family 18 exo-chitinase. Proc Natl Acad Sci USA. 2001;98:8979–84.
Di Rosa M, Malaguarnera G, De Gregorio C, Drago F, Malaguarnera L. Evaluation of CHI3L-1 and CHIT-1 expression in differentiated and polarized macrophages. Inflammation. 2013;36:482–92.
Di Rosa M, Tibullo D, Saccone S, Distefano G, Basile MS, Di Raimondo F, et al. CHI3L1 nuclear localization in monocyte derived dendritic cells. Immunobiology. 2016;221:347–56.
Di Rosa M, Tibullo D, Vecchio M, Nunnari G, Saccone S, Di Raimondo F, et al. Determination of chitinases family during osteoclastogenesis. Bone. 2014;61:55–63.
Tibullo D, Di Rosa M, Giallongo C, La Cava P, Parrinello NL, Romano A, et al. Bortezomib modulates CHIT1 and YKL40 in monocyte-derived osteoclast and in myeloma cells. Front Pharmacol. 2015;6:226.
Qiu QC, Wang L, Jin SS, Liu GF, Liu J, Ma L, et al. CHI3L1 promotes tumor progression by activating TGF-beta signaling pathway in hepatocellular carcinoma. Sci Rep. 2018;8:15029.
Di Rosa M, Malaguarnera L. Chitinase 3 like-1: an emerging molecule involved in diabetes and diabetic complications. Pathobiology. 2016;83:228–42.
Szychlinska MA, Trovato FM, Di Rosa M, Malaguarnera L, Puzzo L, Leonardi R, et al. Co-expression and co-localization of cartilage glycoproteins CHI3L1 and lubricin in osteoarthritic cartilage: morphological, immunohistochemical and gene expression profiles. Int J Mol Sci. 2016;17:359.
Malaguarnera L, Imbesi R, Di Rosa M, Scuto A, Castrogiovanni P, Messina A, et al. Action of prolactin, IFN-gamma, TNF-alpha and LPS on heme oxygenase-1 expression and VEGF release in human monocytes/macrophages. Int Immunopharmacol. 2005;5:1458–69.
Di Rosa M, Sanfilippo C, Libra M, Musumeci G, Malaguarnera L. Different pediatric brain tumors are associated with different gene expression profiling. Acta Histochem. 2015;117:477–85.
Di Rosa M, Szychlinska MA, Tibullo D, Malaguarnera L, Musumeci G. Expression of CHI3L1 and CHIT1 in osteoarthritic rat cartilage model. A morphological study. Eur J Histochem. 2014;58:2423.
Higashiyama M, Tomita K, Sugihara N, Nakashima H, Furuhashi H, Nishikawa M, et al. Chitinase 3-like 1 deficiency ameliorates liver fibrosis by promoting hepatic macrophage apoptosis. Hepatol Res. 2019;49:1316–28.
Kwak EJ, Hong JY, Kim MN, Kim SY, Kim SH, Park CO, et al. Chitinase 3-like 1 drives allergic skin inflammation via Th2 immunity and M2 macrophage activation. Clin Exp Allergy. 2019;49:1464–74.
Sanfilippo C, Nunnari G, Calcagno A, Malaguarnera L, Blennow K, Zetterberg H, et al. The chitinases expression is related to Simian Immunodeficiency Virus Encephalitis (SIVE) and in HIV encephalitis (HIVE). Virus Res. 2017;227:220–30.
Scarpino M, Pinzone MR, Di Rosa M, Madeddu G, Foca E, Martellotta F, et al. Kidney disease in HIV-infected patients. Eur Rev Med Pharmacol Sci. 2013;17:2660–7.
Hall S, Surova Y, Ohrfelt A, Swedish Bio FS, Blennow K, Zetterberg H, et al. Longitudinal measurements of cerebrospinal fluid biomarkers in Parkinson's disease. Mov Disord. 2016;31:898–905.
Sanfilippo C, Longo A, Lazzara F, Cambria D, Distefano G, Palumbo M, et al. CHI3L1 and CHI3L2 overexpression in motor cortex and spinal cord of sALS patients. Mol Cell Neurosci. 2017;85:162–9.
Sanfilippo C, Castrogiovanni P, Imbesi R, Kazakowa M, Musumeci G, Blennow K, et al. Sex difference in CHI3L1 expression levels in human brain aging and in Alzheimer's disease. Brain Res. 2019;1720:146305.
Sanfilippo C, Malaguarnera L, Di Rosa M. Chitinase expression in Alzheimer's disease and non-demented brains regions. J Neurol Sci. 2016;369:242–9.
Mizoguchi E. Chitinase 3-like-1 exacerbates intestinal inflammation by enhancing bacterial adhesion and invasion in colonic epithelial cells. Gastroenterology. 2006;130:398–411.
Deutschmann C, Sowa M, Murugaiyan J, Roesler U, Rober N, Conrad K, et al. Identification of chitinase-3-like protein 1 as a novel neutrophil antigenic target in Crohn's disease. J Crohns Colitis. 2019;13:894–904.
Clough E, Barrett T. The gene expression omnibus database. Methods Mol Biol. 2016;1418:93–110.
Rempel H, Sun B, Calosing C, Pillai SK, Pulliam L. Interferon-alpha drives monocyte gene expression in chronic unsuppressed HIV-1 infection. Aids. 2010;24:1415–23.
Castellanos-Rubio A, Santin I, Martin-Pagola A, Irastorza I, Castano L, Vitoria JC, et al. Long-term and acute effects of gliadin on small intestine of patients on potentially pathogenic networks in celiac disease. Autoimmunity. 2010;43:131–9.
Heap GA, Trynka G, Jansen RC, Bruinenberg M, Swertz MA, Dinesen LC, et al. Complex nature of SNP genotype effects on gene expression in primary human leucocytes. BMC Med Genom. 2009;2:1.
Jorgensen SF, Reims HM, Frydenlund D, Holm K, Paulsen V, Michelsen AE, et al. A cross-sectional study of the prevalence of gastrointestinal symptoms and pathology in patients with common variable immunodeficiency. Am J Gastroenterol. 2016;111:1467–75.
Sangineto M, Graziano G, D'Amore S, Salvia R, Palasciano G, Sabba C, et al. Identification of peculiar gene expression profile in peripheral blood mononuclear cells (PBMC) of celiac patients on gluten free diet. PLoS ONE. 2018;13:e0197915.
Xiao J, Cao H, Chen J. False discovery rate control incorporating phylogenetic tree increases detection power in microbiome-wide multiple testing. Bioinformatics. 2017;33:2873–81.
Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 2004;3:Article3.
Davis S, Meltzer PS. GEOquery: a bridge between the gene expression omnibus (GEO) and BioConductor. Bioinformatics. 2007;23:1846–7.
Fagone P, Nunnari G, Lazzara F, Longo A, Cambria D, Distefano G, et al. Induction of OAS gene family in HIV monocyte infected patients with high and low viral load. Antivir Res. 2016;131:66–73.
Cheadle C, Vawter MP, Freed WJ, Becker KG. Analysis of microarray data using Z score transformation. J Mol Diagn. 2003;5:73–81.
Castrogiovanni P, Li Volti G, Sanfilippo C, Tibullo D, Galvano F, Vecchio M, et al. Fasting and fast food diet play an opposite role in mice brain aging. Mol Neurobiol. 2018;55:6881–933.
Di Rosa M, Malaguarnera L. Genetic variants in candidate genes influencing NAFLD progression. J Mol Med. 2012;90:105–18.
Sanfilippo C, Castrogiovanni P, Imbesi R, Tibullo D, Li Volti G, Barbagallo I, et al. Middle-aged healthy women and Alzheimer's disease patients present an overlapping of brain cell transcriptional profile. Neuroscience. 2019;406:333–44.
Low D, DeGruttola AK, Poltrak A, Mizoguchi A, Mino-Kenudson M, Mizoguchi E. High endogenous expression of chitinase 3-like 1 and excessive epithelial proliferation with colonic tumor formation in MOLF/EiJ mice. PLoS ONE. 2015;10:e0139149.
Low D, Subramaniam R, Lin L, Aomatsu T, Mizoguchi A, Ng A, et al. Chitinase 3-like 1 induces survival and proliferation of intestinal epithelial cells during chronic inflammation and colitis-associated cancer by regulating S100A9. Oncotarget. 2015;6:36535–50.
Pascual V, Dieli-Crimi R, Lopez-Palacios N, Bodas A, Medrano LM, Nunez C. Inflammatory bowel disease and celiac disease: overlaps and differences. World J Gastroenterol. 2014;20:4846–56.
Erzin Y, Uzun H, Karatas A, Celik AF. Serum YKL-40 as a marker of disease activity and stricture formation in patients with Crohn's disease. J Gastroenterol Hepatol. 2008;23:e357–e362362.
Kawada M, Chen CC, Arihiro A, Nagatani K, Watanabe T, Mizoguchi E. Chitinase 3-like-1 enhances bacterial adhesion to colonic epithelial cells through the interaction with bacterial chitin-binding protein. Lab Investig. 2008;88:883–95.
Chen CC, Llado V, Eurich K, Tran HT, Mizoguchi E. Carbohydrate-binding motif in chitinase 3-like 1 (CHI3L1/YKL-40) specifically activates Akt signaling pathway in colonic epithelial cells. Clin Immunol. 2011;140:268–75.
Libreros S, Iragavarapu-Charyulu V. YKL-40/CHI3L1 drives inflammation on the road of tumor progression. J Leukoc Biol. 2015;98:931–6.
Patil NS, Hall FC, Drover S, Spurrell DR, Bos E, Cope AP, et al. Autoantigenic HCgp39 epitopes are presented by the HLA-DM-dependent presentation pathway in human B cells. J Immunol. 2001;166:33–41.
Tang H, Sun Y, Shi Z, Huang H, Fang Z, Chen J, et al. YKL-40 induces IL-8 expression from bronchial epithelium via MAPK (JNK and ERK) and NF-kappaB pathways, causing bronchial smooth muscle proliferation and migration. J Immunol. 2013;190:438–46.
Kawada M, Seno H, Kanda K, Nakanishi Y, Akitake R, Komekado H, et al. Chitinase 3-like 1 promotes macrophage recruitment and angiogenesis in colorectal cancer. Oncogene. 2012;31:3111–233.
Acknowledgements
We would like to show our gratitude to the authors of microarray datasets made available online, for consultation and re-analysis. In addition, I would like to express my gratitude to Oliver Di Rosa, an inspiration in my life.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing financial interests.
Additional information
Responsible Editor: John Di Battista.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Catrogiovanni, P., Musumeci, G., Giunta, S. et al. The expression levels of CHI3L1 and IL15Rα correlate with TGM2 in duodenum biopsies of patients with celiac disease. Inflamm. Res. 69, 925–935 (2020). https://doi.org/10.1007/s00011-020-01371-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00011-020-01371-9