Abstract.
Introduction:
Proteus rods are currently subdivided into five named species, i.e. Proteus mirabilis, P. vulgaris, P. penneri, P. hauseri, and P. myxofaciens, and three unnamed Proteus genomospecies 4 to 6. Based on the serospecificity of the lipopolysaccharide (LPS; O-antigen), strains of P. mirabilis and P. vulgaris were divided into 49 O-serogroups and 11 additional O-serogroups were proposed later. About 15 further O-serogroups have been proposed for the third medically important species, P. penneri. Here the serological classification of P. vulgaris strain TG 251, which does not belong to these serogroups, is reported. Serological investigations also allowed characterization of the epitope specificity of its LPS.
Materials and Methods:
Purified LPSs from five Proteus strains were used as antigens in enzyme immunosorbent assay (EIA), SDS/PAGE, and Western blot and alkali-treated LPSs in the passive immunohemolysis (PIH) test, inhibition of PIH and EIA, and absorption of the rabbit polyclonal O-antisera with the respective LPS.
Results:
The serological studies of P. vulgaris TG 251 LPS indicated the identity of its O-polysaccharide with that of P. penneri O65. The antibody specificities of P. vulgaris TG 251 and P. penneri O65 O-antisera, were described.
Conclusions:
P. vulgaris TG 251 was classified to the Proteus O65 serogroup. Two disaccharide-associated epitopes present in P. vulgaris TG 251 and P. penneri O65 LPSs are suggested to be responsible for cross-reactions with three heterologous Proteus strains.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Much has been written about the taxonomy of Proteus since the original publication by Hauser, who established the genus, in 1885 [2]. Currently, the genus Proteus consists of five named species (P. mirabilis, P. vulgaris, P. penneri, P. myxofaciens, and P. hauseri) and three unnamed Proteus genomospecies 4, 5 and 6 [6, 7]. Proteus rods are widespread in the environment and make up part of the normal flora of the human gastrointestinal tract. Bacteria of the genus Proteus are the third pathogen (after Escherichia and Klebsiella) which cause uncomplicated cystis, pyelonephritis, and prostatitis, particularly in hospital-acquired cases [12]. It was also suggested that P. mirabilis might play an etiopathogenic role in rheumatoid arthritis [16].
The serological specificity of Proteus strains is defined by the chemical structure of the O-specific polysaccharide chain (O-antigen) of the lipopolysaccharide (LPS, endotoxin). According to the serological specificity of the O antigens, strains of two species, P. mirabilis and P. vulgaris, were classified into 60 O-serogroups [5, 8], including 49 numbered serogroups (O1–O49) [5]. Fifteen further O-serogroups were proposed for the strains of P. penneri [3, 10, 17]. The aim of the present study was a serological classification of Proteus vulgaris strain TG 251, which does not belong to the Kauffmann-Perch scheme [5, 8]. Using serological and structural data, the epitopes of P. vulgaris TG 251 LPS were investigated.
Materials and Methods
Bacterial strains
Proteus vulgaris strain TG 251 was kindly provided by Prof. J. L. Penner (Department of Medical Genetics, University of Toronto, Toronto, Ontario, Canada). Thirty-nine P. mirabilis and 27 P. vulgaris strains were from the Czech National Collection of Type Cultures (National Institute of Public Health, Prague, Czech Republic). Twenty-four P. penneri, one Proteus genomospecies, and one P. hauseri strain were kindly provided by D. J. Brenner and C. M. O’Hara (Centers for Disease Control and Prevention, Atlanta, Georgia, USA). A P. myxofaciens (CCUG 18769) strain was received from Dr. E. Falsen (Cultures Collection, University of Goeteborg (CCUG), Goeteborg, Sweden).
Cultivation of bacteria, isolation and saponification of the LPSs
Bacteria were cultivated under aerobic conditions in nutrient broth (BTL, Łódź, Poland). LPSs were obtained by the extraction of dry bacterial mass with hot phenol/water [15] and purified with aqueous 50% trichloracetic acid at 4°C, followed by dialysis of the supernatant [19]. Alkali-treated LPSs were prepared by saponification of the LPSs with 0.25 M NaOH (56°C, 2 h), followed by precipitation with ethanol. The purified LPSs were used as antigens in the enzyme immunosorbent assay (EIA), SDS/polyacrylamide gel electrophoresis (SDS/PAGE), and Western blotting, and the alkalitreated LPSs in the passive immunohemolysis (PIH) test, inhibition of EIA and PIH, and absorption of the antisera with the respective LPSs.
Preparation of the O-polysaccharide
Delipidation of the P. vulgaris TG 251 LPS was performed by mild acid degradation with aqueous 2% HOAc at 100°C until precipitation of lipid A. The precipitate was removed by centrifugation (13,000×g, 20 min) and the supernatant was fractionated by gel-permeation chromatography on a column (56×2.6 cm) of Sephadex G-50 (S) (Amersham Biosciences, Sweden) in 0.05 M pyridinum acetate buffer (pH 4.5), monitored using a differential refractometer (Knauer, Germany). The yield of the high-molecular-mass O-specific polysaccharide of P. vulgaris TG 251 was 19% of the LPS weight.
NMR spectroscopy
The sample was freeze-dried twice from D2O prior to measurement. 13C-NMR spectrum was recorded with a Bruker DRX-500 spectrometer (Germany) for a solution in D2O at 50°C using internal acetone (δC: 31.45) as reference. Standard Bruker software (XWINNMR 2.6) was used to acquire and process the NMR data.
Rabbit antisera and serological assays
The polyclonal O-antisera were obtained by immunization of New Zealand white rabbits with heat-inactivated bacteria of P. vulgaris TG 251 and P. penneri O65 according to the published procedure [18]. All serological tests were performed according to procedures described in detail elsewhere [11].
Results and Discussion
In this paper we present the results of serological and immunochemical studies of LPS isolated from a Proteus vulgaris strain described in the Penner collection as TG 251 [8]. As judged by the essential identity of the 13C-NMR spectra (Fig. 1), the O-specific polysaccharide isolated from the P. vulgaris TG 251 LPS has the same repeating unit as that of Proteus penneri 34 from serogroup O65, whose structure we determined earlier [14, 17] (Fig. 2). Therefore, in the serological investigations performed in this study, a second O-antiserum, that against P. penneri O65, was included. The LPSs of a number of Proteus strains with known O-polysaccharide structure were tested with polyclonal rabbit O-antisera against P. vulgaris TG 251 and P. penneri O65 by PIH and EIA to reveal possible serological relatedness of these bacteria.
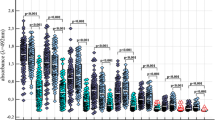
Of 93 tested Proteus LPSs, including those of 39 P. mirabilis, 27 P. vulgaris, 24 P. penneri, one P. hauseri, one P. myxofaciens, and one Proteus genomospecies, only four strains were cross-reactive with P. vulgaris TG 251 O-antiserum, namely the LPSs of P. mirabilis O16, O34, and O50 and P. penneri O65. The strongest cross-reactivity, with P. penneri O65 LPS, was on the same level as with the homologous P. vulgaris LPS, thus suggesting again the close O-antigenic similarity of both strains. Based on this finding, P. penneri O65 O-antiserum was included in the subsequent detailed serological investigation, which involved PIH and EIA tests, inhibition of reactions in these tests, and absorption of O-antisera with the cross-reactive LPSs, followed by repeated PIH as well as Western blotting (Table 1, Fig. 3).
In PIH and EIA, both O-antisera reacted equally strongly with the homologous LPSs. A weaker reaction was observed with the heterologous P. mirabilis O16, O34, and O50 LPSs. Accordingly, the homologous LPSs were the strongest inhibitors of the reaction in both PIH and EIA, whereas the heterologous LPSs showed a weaker inhibitory activity. It is worth mentioning that P. penneri O65 O-antiserum cross-reacted more weakly with all heterologous LPSs in comparison with P. vulgaris TG 251 O-antiserum.
In Western blot, both P. vulgaris TG 251 and P. penneri O65 O-antisera clearly recognized the same high-molecular-mass O-polysaccharide containing LPS species of both homologous strains, but the binding patterns of their low-molecular-mass LPS species, restricted to the core-lipid A moiety, were different (Fig. 3 A, B). All heterologous LPSs reacted significantly more weakly and bound only to high-molecular-mass LPS species.
The data obtained for the LPSs from P. vulgaris TG 251 and P. penneri O65 were essentially identical in all tests used. These results were in agreement with the structural investigation data of the P. vulgaris TG 251 O-specific polysaccharide, which revealed the same repeating unit as that of P. penneri O65, described in literature [14, 17]. They also support Penner’s and Hennessy’s conclusion [8] that P. vulgaris TG 251 does not belong to the Kauffmann and Perch scheme of the serological classification of P. mirabilis and P. vulgaris strains (which also does not include P. penneri strains). Therefore, we propose classifying the strain studied, P. vulgaris TG 251, to the Proteus O65 serogroup, in which P. penneri 34 (O65) has until now been the only representative.
Epitope characterization of P. vulgaris TG 251 LPS
In order to determine epitope specificity, the LPSs studied were tested using PIH with P. vulgaris TG 251 and P. penneri O65 O-antisera (Table 2). The reactivity of all the tested antigens was completely abolished when both O-antisera were absorbed with the homologous LPSs (Table 2). However, this absorption leaves a small fraction of antibodies in both O-antisera cross-reacting (1:1600) with the homologous LPSs of P. vulgaris TG 251 and P. penneri O65. These results could be accounted for by the difference in the LPS core region of the low-molecular-mass LPS species of these two strains shown in Western blot (Fig. 3). Absorption of both O-antisera with LPSs of P. mirabilis O16 and O34 removed antibodies cross-reacting to each other and decreased the reaction with the homologous antigens, but did not influence the reactivity with P. mirabilis O50 LPS.
A comparison of these serological results with the chemical structures of the O-specific polysaccharides of the cross-reacting LPSs described in literature [4, 9, 13] suggests that, in addition to the LPS core-specific antibodies, in both P. vulgaris TG 251 and P. penneri O65 O-antisera there are two more, major types of antibodies which recognize distinct epitopes in common with the LPS studied (Fig. 2). The first common epitope on the O-specific polysaccharides of P. mirabilis O16 and O34, responsible for their serological cross-reactions with both O-antisera, could be the β-D-GalpNAc-(1→4)-β-D-GalpNAc disaccharide fragment. This conclusion was confirmed by an absorption experiment (Table 2). Absorption with either of the heterologous LPSs (P. mirabilis O16 or O34) removed from both investigated O-antisera those antibodies which recognize the other strain from the pair and vice versa. Another independent fraction of antibodies in the investigated O-antisera seems to recognize the second common epitope associated with the β-D-GalpNAc-(1→4)-β-D-Galp disaccharide fragment shared by the homologous O-specific polysaccharides (those of P. vulgaris TG 251 and P. penneri O65) and that of P. mirabilis O50 (Fig. 2).
In conclusion, the serological results obtained in this study, combined with structural analysis data, demonstrated the identity of the O-specific polysaccharides from P. vulgaris TG 251 and P. penneri 34 (O65) LPSs and allowed classification of the former to the Proteus O65 serogroup, formerly represented by the single strain P. penneri 34. In O-antisera against both serogroup O65 strains studied, three different fractions of antibodies were detected and their LPS epitopes could be tentatively determined as follows. Major antibodies of the first type recognize the β-D-GalpNAc-(1→4)-β-D-GalpNAc disaccharide as a common fragment present in the O-specific polysaccharides of P. vulgaris TG 251, P. penneri O65, and P. mirabilis O16 and O34 LPSs. Major antibodies of the second type bind to the β-D-GalpNAc(1→4)-β-D-Galp disaccharide fragment shared by the O-specific polysaccharides of P. vulgaris TG 251, P. penneri O65, and P. mirabilis O55 LPSs. Antibodies of the third type react with epitope(s) localized on the LPS core of P. vulgaris TG 251 and P. penneri O65 LPSs, whose nature as well as whole core structures remain unknown.
Abbreviations
- EIA:
-
enzyme immunosorbent assay
- LPS:
-
lipopolysaccharide
- PAGE:
-
polyacrylamide gel electrophoresis
- PIH:
-
passive immunohemolysis
References
Drzewiecka D., Zych K. and Sidorczyk Z. (2004): Characterization and serological classification of a collection of Proteus penneri clinical strains. Arch. Immunol. Ther. Exp., 52, 121–128.
Hauser G. (1885): Über Fäulnissbacterien und deren Beziehungen zur Septicämie. Ein Beitrag zur Morphologie der Spaltpilze, Vogel, Leipzig, Germany, p. 12
Knirel Y. A., Kaca W., Różalski A. and Sidorczyk Z. (1999): Structures of O-antigenic polysaccharides of Proteus bacteria. Pol. J. Chem., 73, 895–907.
Kołodziejska K., Kondakova A. N., Zych K., Senchenkova S. N., Shashkov A. S., Knirel Y. A. and Sidorczyk Z. (2003): Structure of the O-polysaccharide of a serologically separate strain of Proteus mirabilis TG 332, from a new proposed Proteus serogroup O50. Carbohydr. Res., 338, 2105–2109.
Larsson P. (1984): Serology of Proteus mirabilis and Proteus vulgaris. Methods Microbiol., 14, 187–214.
O’Hara C. M., Brenner F. W. and Miller J. M. (2000): Classification, identification, and clinical significance of Proteus, Providencia, and Morganella. Clin. Microbiol. Rev., 13, 534–546.
O’Hara C. M., Brenner F. W., Steigewalt A. G., Hill B. C., Holmes B., Grimont P. A., Hawkey P. M., Penner J. L., Miller J. M. and Brenner D. J. (2000): Classification of Proteus vulgaris biogroup 3 with recognition of Proteus hauseri sp. nov. nom. rev. and unnamed Proteus genomospecies 4, 5 and 6. Int. J. Syst. Evol. Microbiol., 50, 1869–1875.
Penner J. L. and Hennessy J. N. (1980): Separate O-grouping schemes for serotyping clinical isolates of Proteus vulgaris and Proteus mirabilis. J. Clin. Microbiol., 12, 304–309.
Perepelov A. V., Kołodziejska K., Kondakova A. N., Wykrota M., Knirel Y. A., Sidorczyk Z and Różalski A. (2004): Structure of the O-polysaccharide of Proteus serogroup O34 containing 2-acetamido-2-deoxy-alpha-D-galactosyl phosphate. Carbohydr. Res., 339, 2145–2149.
Sidorczyk Z., Zych K., Kołodziejska K., Drzewiecka D. and Zabłotni A. (2002): Progress in serological classification of further strains from genus Proteus and determination of epitopes and new serogroups. Second German-Polish-Russian Meeting on Bacterial Carbohydrates, Moscow, September 10–12, 2002.
Sidorczyk Z., Zych K., Toukach F. V., Arbatsky N. P., Zabłotni A., Shashkov A. S. and Knirel Y. A. (2002): Structure of O-polysaccharide and classification of Proteus mirabilis strain G1 in Proteus serogroup O3. Eur. J. Biochem., 269, 1406–1412.
Stamm W. E. (1999): Urinary tract infection. In Root R. K. (ed.): Clinical infection diseases: a practical approach. Oxford University Press, Oxford, UK, pp. 549–656.
Toukach F. V., Arbatsky N. P., Shashkov A. S., Knirel Y. A., Zych K. and Sidorczyk Z. (2001): Structure of the O-specific polysaccharide of Proteus mirabilis O16 containing ethanolamine phosphate and ribitol phosphate. Carbohydr. Res., 331, 213–218.
Toukach F. V., Shashkov A. S., Arbatsky N. P., Knirel Y. A., Zych K. and Sidorczyk Z. (1998): Structure of the O-specific polysaccharide of Proteus penneri 34. Carbohydr. Res., 312, 97–101.
Westphal O. and Jann K. (1965): Bacterial lipopolysaccharides. Extraction with phenol/water and further application of the procedure. Methods Carbohydr. Chem., 5, 83–91.
Wilson C., Thakore A., Isenberg D. and Ebringer A. (1997): Correlation between anti-Proteus antibodies and isolation rates of P. mirabilis in rheumatoid arthritis. Rheumatol. Int., 16, 187–189.
Zych K., Kowalczyk M., Knirel Y. A. and Sidorczyk Z. (2000): New serogroups of the genus Proteus consisting of Proteus penneri strains only. Determination of some LPS epitopes responsible for specificity. Adv. Exp. Med. Biol., 485, 339–344.
Zych K., Świerzko A. and Sidorczyk Z. (1992): Serological characterization of Proteus penneri species novum. Arch. Immunol. Ther. Exp., 40, 89–92.
Zych K., Toukach F. V., Arbatsky N. P., Kołodziejska K., Senchenkova S. N., Shashkov A. S., Knirel Y. A. and Sidorczyk Z. (2001): Structure of the O-specific polysaccharide of Proteus mirabilis D52 and typing of this strain to Proteus serogroup O33. Eur. J. Biochem., 268, 4346–4351.
Acknowledgment
This work was supported by the State Committee for Scientific Research (KBN, Poland, grant no. 2 PO5A 085 26). The authors thank Janusz Włodarczyk for excellent technical assistance in the preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Zych, K., Kołodziejska, K., Drzewiecka, D. et al. Serological classification and epitope specificity of Proteus vulgaris TG 251 from Proteus serogroup O65. Arch. Immunol. Ther. Exp. 55, 187–191 (2007). https://doi.org/10.1007/s00005-007-0020-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00005-007-0020-z