Abstract
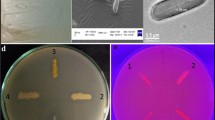
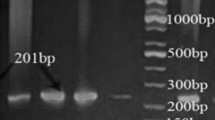
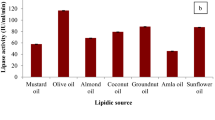
The study was done to isolate, identify, and characterize a good lipolytic strain from soil. Lipolytic strain isolation was done using tributyrin agar medium. The biochemical testing and 16S rRNA gene sequencing analysis was done for identification. The enzyme was purified using ammonium sulfate precipitation and column chromatography. Results have shown a novel high lipolytic strain of P. aeruginosa JCM5962(T), isolated from soil of sugarcane field. The 16S rRNA sequence analysis confirmed the strain as P. aeruginosa JCM5962(T); further, the sequence was submitted to Genbank (KX946966.1). The isolate produced an extracellular lipase which was purified as single band of 31 kDa. Maximum lipase activity was observed at 50 °C and pH 8.0. Activity was enhanced in the presence of cobalt and benzene solvent, whereas mercury, sodium dodecyl sulfate, and chloroform inhibited it. The enzyme’s marked stability and activity at high temperature, alkaline pH and organic solvents suggest that this can be effectively used in a variety of applications in industries and as biotechnological tools.






Similar content being viewed by others
References
Ahmed EH, Raghavendra T, Madamwar D (2010) A thermostable alkaline lipase from a local isolate Bacillus subtilis EH 37: characterization, partial purification and application in organic synthesis. Appl Biochem Biotechnol 160(7):2102–2113
Anbu P, Hur BK (2014) Isolation of an organic solvent-tolerant bacterium Bacillus licheniformis PAL05 that is able to secrete solvent-stable lipase. Biotechnol Appl Biochem 61(5):528–534
Arpigny JL, Jaeger KE (1999) Bacterial lipolytic enzymes: classification and properties. Biochem J 343:177–183
Bisht D, Yadav SK, Gautam P, Darmwal NS (2013) Simultaneous production of alkaline lipase and protease by antibiotic and heavy metal tolerant Pseudomonas aeruginosa. J Basic Microbiol 53(9):715–722
Bisht D, Yadav SK, Darmwal NS (2014) An oxidant and organic solvent tolerant alkaline lipase by P. aeruginosa mutant: downstream processing and biochemical characterization. Braz J Microbiol 44(4):1305–1314
Boonsinthai B, Phutraku S (1999) Effect of metal ions, inhibitors and denaturants on extracellular lipases from three thermophile isolates and their clones. Chiang Mai J Sci 26(1):25–43
Collee JG, Fraser AG, Marmion BP, Simmons A, Mackie MC (1996) Practical medical microbiology, 14th edn. Churchill Livingstone, London
Davis BJ (1964) Disc electrophoresis II. Method and application to human serum proteins. Ann N Y Acad Sci 121:321–349
Davranov K (1994) Microbial lipases in biotechnology (review). Appl Biochem Microbiol 30:427–432
Demain AL (2000) Microbial biotechnology. Trends Biotechnol 18:26–31
Ghori MI, Iqbal MJ, Hameed A (2011) Characterization of a novel lipase from Bacillus sp. isolated from tannery wastes. Braz J Microbiol 42(1):22–29
Gokbulut AA, Arslanoglu A (2013) Purification and biochemical characterization of an extracellular lipase from psychrotolerant Pseudomonas fluorescens KE38. Turk J Biol 37:538–546
Guncheva M, Zhiryakova D (2011) Catalytic properties and potential applications of Bacillus lipases. J Mol Catal B Enzym 68(1):1–21
Guncheva M, Tashev E, Zhiryakova D, Tosheva T, Tzokova N (2011) Immobilization of lipase from Candida rugosa on novel phosphorous-containing polyurethanes: application in wax ester synthesis. Process Biochem 46(4):923–930
Gupta R, Gupta N, Rathi P (2004) Bacterial lipases: an overview of production, purification and biotechnological properties. Appl Microbiol Biotechnol 64:763–781
Gupta A, Joseph B, Mani A, Thomas G (2008) Biosynthesis and properties of an extracellular thermostable serine alkaline protease from Virgibacillus pantothenticus. World J Microbiol Biotechnol 24:237–243
Hasan F, Shah AA, Hameed A (2006) Industrial applications of microbial lipases. Enzym Microb Technol 39:235–251
Jaeger KE, Eggert T (2002) Lipases for biotechnology. Curr Opin Biotechnol 13(4):390–397
Ji Q, Xiao S, He B, Liu X (2010) Purification and characterization of an organic solvent-tolerant lipase from Pseudomonas aeruginosa LX1 and its application for biodiesel production. J Mol Catal B Enzym 66:264–269
Kakugawa K, Shobayashi M, Suzuki O, Miyakawa T (2002) Purification and characterization of a lipase from the glycolipid-producing yeast Kurtzmanomyces sp. I-11. Biosci Biotechnol Biochem 66(5):978–985
Kanderi DK, Yadav U, Satyanarayana SV, Verma S (2014) Characterization of partially purified lipase from Saccharomyces cerevisiae. Int J Pharm Pharm Sci 6(8):514–517
Karadzic I, Masui A, Zivkovic LI, Fujiwara N (2006) Purification and characterization of an alkaline lipase from Pseudomonas aeruginosa isolated from putrid mineral cutting oil as component of metal working fluid. J Biosci Bioeng 102(2):82–89
Kumar A, Dhar K, Kanwar SS, Arora PK (2016) Lipase catalysis in organic solvents: advantages and applications. Biol Proced Online 18(2):1–11
Martinez A, Soberon CG (2001) Characterization of the lipA gene encoding the major lipase from Pseudomonas aeruginosa strain IGB83. Appl Microbiol Biotechnol 56:731–735
Mukke VK, Chinte DN (2012) Impact of heavy metal induced alterations in lipase activity of fresh water crab, Barytelphusa guerini. J Chem Pharm Res 4(5):2763–2766
Nielsen PM, Brask J, Fjerbaek L (2008) Enzymatic biodiesel production: technical and economical considerations. Eur J Lipid Sci Technol 110:692–700
Paje ML, Neilan BA, Couperwhite I (1997) A Rhodococcus species that thrives on medium saturated with liquid benzene. Microbiol 143:2975–2981
Pourahmad JR, Ebadi R (2013) Study the expression of mara gene in ciprofloxacin and tetracycline resistant mutants of Esherichia coli. Iran J Pharm Res 12:923–928
Pramanik K, Saren S, Mitra S, Ghosh PK, Maiti TK (2018) Computational elucidation of phylogenetic, structural and functional characteristics of Pseudomonas lipases. Comput Biol Chem 74:190–200
Reetz MT (2002) Lipases as practical biocatalysts. Curr Opin Chem Biol 6(2):145–150
Romero CM, Pera LM, Loto F, Vallejos C, Castro G, Baigori M (2012) Purification of an organic solvent-tolerant lipase from Aspergillus niger MYA 135 and its application in ester synthesis. Biocatal Agric Biotechnol 1(1):25–31
Sachan S, Singh A (2015) Lipase enzyme and its diverse role in food processing industry. Everyman’s Sci 4:214–218
Sachan S, Chandra VY, Yadu A, Singh A (2017) Cobalt has enhancing effect on extracellular lipases isolated from Pseudomonas aeruginosa JCM5962(T). Int J PharmTech Res 10(1):45–49
Saranya P, Kumari HS, Rao BP, Sekaran G (2014) Lipase production from a novel thermo-tolerant and extreme acidophile Bacillus pumilus using palm oil as the substrate and treatment of palm oil-containing wastewater. Environ Sci Pollut Res 21:3907–3919
Schmid RD, Verger R (1998) Lipases: interfacial enzymes with attractive applications. Angew Chem Int Ed Engl 37(12):1608–1633
Shah KR, Bhatt SA (2011) Purification and characterization of lipase from Bacillus subtilis Pa2. J Biochem Tech 3(3):292–295
Siddiqui KS, Azhar MJ, Rashid MH, Rajoka MI (1997) Stability and identification of active-site residues of carboxym ethyl cellulases from Aspergillus niger and Cellulomonas biazotea. Folia Microbiol (Praha) 42:312–318
Stuer W, Jaeger KE, Winkler U (1986) Purification of extracellular lipase from Pseudomonas aeruginosa. J Bacteriol 168:1070–1074
Svendsen A, Borch K, Barfoed M, Nielsen T, Gormsen E, Patkar S (1995) Biochemical properties of cloned lipases from Pseudomonas family. Biochim Biophys Acta 1259:9–17
Thakur S (2012) Lipases, its sources, properties and applications: a review. Int J Sci Eng Res 3(7):1–29
Tiwari P, Upadhyay MK, Silawat N, Verma HN (2011) Optimization and characterization of a thermo tolerant lipase from Cryptococcus albidus. Der Pharma Chemica 3(4):501–508
Torres S, Castro GR (2003) Organic solvent resistant lipase produced by thermoresistant bacteria. New Horizons Biotechnol, Netherlands: Springer 18(2):113–122
Ulker S, Ozel A, Colak A, Karaoglu SA (2011) Isolation, production, and characterization of an extracellular lipase from Trichoderma harzianum isolated from soil. Turk J Biol 35(5):543–550
Verma ML, Azmi W, Kanwar SS (2008) Microbial lipases: at the interface of aqueous and non-aqueous media: a review. Acta Microbiol Immunol Hung 55(3):265–294
Zouaoui B, Bouziane A, Ghalem BR (2012) Isolation, purification and properties of lipase from Pseudomonas aeruginosa. Afr J Biotechnol 11(60):12415–12421
Acknowledgements
The authors are thankful to Chairperson, Pro VC, Amity University Lucknow campus and Head, Amity Institute of Biotechnology for providing support and all necessary facilities to conduct the work. Authors are also thankful to IMTECH, Chandigarh, for 16S rRNA sequencing.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sachan, S., Iqbal, M.S. & Singh, A. Extracellular lipase from Pseudomonas aeruginosa JCM5962(T): Isolation, identification, and characterization. Int Microbiol 21, 197–205 (2018). https://doi.org/10.1007/s10123-018-0016-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10123-018-0016-z