Abstract
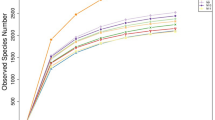
Bacterial communities in buffalo rumen were characterized using a culture-independent approach for a pooled sample of rumen fluid from 3 adult Surti buffaloes. Buffalo rumen is likely to include species of various bacterial phyla, so 16S rDNA sequences were amplified and cloned from the sample. A total of 191 clones were sequenced and similarities to known 16S rDNA sequences were examined. About 62.82% sequences (120 clones) had >90% similarity to the 16S rDNA database sequences. Furthermore, about 34.03% of the sequences (65 clones) were 85–89% similar to 16S rDNA database sequences. For the remaining 3.14%, the similarity was lower than 85%. Phylogenetic analyses were also used to infer the makeup of bacterial communities in the rumen of Surti buffalo. As a result, we distinguished 42 operational taxonomic units (OTUs) based on unique 16S r DNA sequences: 19 OTUs affiliated to an unidentified group (45.23% of total OTUs), 11 OTUs of the phylum Firmicutes, also known as the low G+C group (26.19%), 7 OTUs of theCytophaga-Flexibacter-Bacteroides phylum (16.66%), 4 OTUs of Spirochaetes (9.52%), and 1 OTU of Actinobacteria (2.38%). These include 10 single-clone OTUs, so Good’s coverage (94.76%) of 16S rRNA libraries indicated that sequences identified in the libraries represent the majority of bacterial diversity present in rumen.
Similar content being viewed by others
References
An D, Dong X, Dong Z, 2005. Prokaryote diversity in the rumen of yak (Bos grunniens) and Jinnan cattle (Bos taurus) estimated by 16S rDNA homology analyses. Anaerobe 11: 207–221.
Acinas SG, Sarma-Rupavtarm R, Klepac-Ceraj V, Polz MF, 2005. PCR-induced sequence artifacts and bias: insights from comparison of two 16S rRNA clone libraries constructed from the same sample. Appl Environ Microbiol 71: 8966–8969.
Buckley DH, Schmidt TM, 2003. Diversity and dynamics of microbial communities in soils from agro-ecosystems. Environ Microbiol 5: 441–452.
Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostelland J, Wheeler D, 2007. GenBank Nucleic Acids Res (35) D1-D25.
Brulc M, Antonopoulos D, Millera ME, Wilsona B, Anthony MK, et al. 2009. Gene-centric metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases. Proc Natl Acad Sci USA 6: 1948–1953.
Dehority BA, Tirabasso PA, Grifo AP, 1989. Most-probable-number procedures for enumerating ruminal bacteria, including the simultaneous estimation of total and cellulolytic numbers in one medium. Appl Environ Microbiol 55: 2789–2792.
Deng W, Wanapat M, Chen S, Ma J, Dongmei Xi, et al. 2007. Phylogenetic analysis of 16S rDNA sequences manifest rumen bacterial diversity in gayals (Bos frontalis) fed fresh bamboo leaves and twigs (Sinarumdinaria). Asian-Aust. J Animal Sci 7(20): 1057–1066.
Dowd SE, Callaway TR, Wolcott RD, Sun Y, McKeehan T, et al. 2008. Evaluation of the bacterial diversity in the feces of cattle using 16SrDNAbacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiology 8:125.
Edwards JE, McEwan NR, Travis AJ, Wallace RJ, 2004. 16S rDNA library-based analysis of ruminal bacterial diversity. Antonie Van Leeuwenhoek 86: 263–281.
Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, et al. 2005. Diversity of the human intestinal microbial flora. Science 308: 1635–1638.
Felsenstein J, 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evol 39: 783–791.
Hungate RE, 1969. Aroll tube method for cultivation of strict anaerobes. In: Norris JR, Ribbons DW, eds.Methods in microbiology, Vol. 3B. London and New York: Academic Press: 117–132.
ICAR, 1998. Nutrient requirements of livestock and poultry. Indian Council of Agricultural Research, New Delhi, India.
Khampa S, Wanapat M, Wachirapakorn C, Nontaso N, Wattiaux M, 2006. Effects of urea level and sodium DL-malate in concentrate containing high cassava chip on ruminal fermentation efficiency, microbial protein synthesis in lactating dairy cows raised under tropical conditions. Asian-Aust J Anim Sci 19: 837–844.
Koike S, Yoshitani S, Kobayashi Y, Tanaka K, 2003. Phylogenetic analysis of fiber-associated rumen bacterial community and PCR detection of uncultured bacteria. FEMS Microbiol Lett 229: 23–30.
Krause DO, Russell JB, 1996. How many ruminal bacteria are there? J Dairy Sci 79: 1467–1475.
Krause DO, Denman SE, Mackie RI, Morrison M, Rae AL, et al. 2003. Opportunities to improve fibre degradation in the rumen: microbiology, ecology, and genomics. FEMS Microbiol Rev 27: 663–693.
Kumar S, Nagarajan M, Sandhu J, Kumar N, Behl V, 2007. Phylogeography and domestication of Indian river buffalo. BMC Evol Biol 7:186.
Lane DJ, 1991. 16S/23S rRNA sequencing. In: Stackebrandt E and Goodfellow M, eds. Nucleic acid techniques in bacterial systematic. New York: John Wiley & Sons: 115–175.
Latham MJ, Storry JE, Sharpe ME, 1972. Effect of low roughage diets on the microflora and lipid metabolism in the rumen. Appl Microbiol 24: 871–877.
Madden TL, Tatusov RL, Zhan J, 1996. Application of network BLAST server. Meth Enzymol 266: 131–141.
Maidak BL, Cole JR, Lilburn TG, Jr CTP, Saxman PR, Farris RJ, et al. 2001. The RDP-II (Ribosomal Database Project). Nucl Acids Res 29: 173–174.
Miron J, BenGhedalia D, Morrison M, 2001. Invited review: adhesion mechanisms of rumen cellulolytic bacteria. J Dairy Sci 84: 1294–1309.
Schloss PD, Handelsman J, 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol 71: 1501–1506.
Shin EC, Cho KM, Lim WJ, Hong SY, An CL, et al. 2004. Phylogenetic analysis of protozoa in the rumen contents of cow based on the 18S rDNA sequences. J Appl Microbiol 97: 378–383.
Srinivas B, Krishnamurthy U, 2005. Influence of diet-induced changes in rumen microbial characteristics on gas production kinetics of straw substratesin vitro. Asian-Aust J Anim Sci 18: 990–996.
Stewart CS, Flint HJ, Bryant MP, 1997. The rumen bacteria. In: Hobson PN, Stewart CS, eds.The rumen microbial ecosystem. 2nd ed. New York: Chapman and Hall: 10–72.
Sylvester JT, Karnati SKR, Yu Z, Morrison M, Firkins JL, 2004. Development of an assay to quantify rumen ciliate protozoal biomass in cows using real-time PCR. J Nutr 134: 3378–3384.
Tajima K, Aminov RI, Nagamine T, Ogata K, Nakamura M, et al. 1999. Rumen bacterial diversity as determined by sequence analysis of 16S rDNA libraries. FEMS Microbiol Ecol 29: 159–169.
Tajima K, Aminov RI, Nagamine T, Matsui H, et al. 2001. Diet-dependent shifts in the bacterial population of the rumen revealed with real-time PCR. Appl Environ Microbiol 67: 2766–2774.
Tamura K, Dudley J, Nei M, Kumar S, 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution 24:1596–1599.
Thompson JD, Higgins DG, Gibson TJ, 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res 22: 4673–4680.
Vandamme P, Pot B, Gillis M, Vos De P, et al. 1996. Polyphasic Taxonomy, a consensus approach to bacterial systematics. Microbiol Rev 60: 407–438.
Whitford MF, Forster RJ, Beard CE, Gong J, Teather RM, 1998. Phylogenetic analysis of rumen bacteria by comparative sequence analysis of cloned 16S rRNA genes. Anaerobe 4: 153–163.
Wintzingerode Von F, Goebel UB, Stackebrandt E, 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev 21: 213–229.
Woese CR, Kandler O, Wheelis ML, 1990. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria and Eucarya. Proc Natl Acad Sci USA 87: 4576–4579.
Yuhei O, Hidenori H, Mitsuo S, Hisao I, Yoshimi B, 2005. Culture-independent analysis of fecal microbiota in cattle. Biosci Botechnol Bochem 69: 1793–1797.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Pandya, P.R., Singh, K.M., Parnerkar, S. et al. Bacterial diversity in the rumen of Indian Surti buffalo (Bubalus bubalis), assessed by 16S rDNA analysis. J Appl Genet 51, 395–402 (2010). https://doi.org/10.1007/BF03208869
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF03208869