Abstract
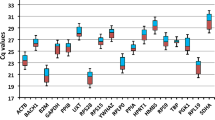
Expression patterns of candidate genes with important functions in animal metabolism can help to identify potential molecular markers for cattle production traits. Reverse transcription followed by polymerase chain reaction is a method for rapid and accurate mRNA quantification. However, for exact comparison of mRNA quantity in various samples or tissues, it is important to choose appropriate reference genes. In cattle, little information is available on the expression stability of housekeeping genes (HKGs). The aim of the present study is to develop a set of reference genes that can be used for normalization of concentrations of mRNAs of genes expressed in the bovine liver, kidney, pituitary and thyroid. The study was performed on 6-, 9-, and 12-month-old bulls of dairy and meat cattle breeds. Six HKGs were investigated:ACTB, GAPDH, HPRTI, SDHA, TBP, andYWHAZ. The most stably expressed potential reference HKGs differed among tissues/organs examined:ACTB, TBP, YWHAZ, GAPDH, HPR TI, andSDHA in the liver;GAPDH andYWHAZ in the kidney;GAPDH andSDHA in the pituitary; andTBP andHPRTI in the thyroid. The results showed that the use of a single gene fornormalization may lead to relatively large errors, so it is important to use multiple control genes based on a survey of potential reference genes applied to representative samples from specific experimental conditions.
Similar content being viewed by others
References
Bionaz M, Loor JJ, 2007. Identification of reference genes for quantitative real-time PCR in the bovine mammary gland during the lactation cycle. Physiol Genomics 29: 312–319.
Blanquicett C, Johnson MR, Heslin M, Diasio RB, 2002. Housekeeping gene variability in normal and carcinomatous colorectal and liver tissues: applications in pharmacogenomic gene expression studies. 303: 209–214.
Bustin SA, 2002. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol 29: 23–39.
De Ketelaere A, Goossens K, Peelman L, Burvenich C. 2006. Technical note: validation of internal control genes for gene expression analysis in bovine polymorphonuclear leukocytes. J Dairy Sci 89: 4066–4069.
Deindl E, Boengler K, van Royen N, Schaper W, 2002. Differential expression ofGAPDH and beta3-actin in growing collateral arteries. Mol Cell Biochem 236: 139–146.
Erkens T, Van Poucke M, Vandesompele J, Goossens K, Van Zeveren A, Peelman LJ, 2006. Development of a new set of reference genes for normalization of real-time RT-PCR data of porcine backfat and longissimus dorsi muscle, and evaluation with PPARGC1A. BMC Biotechnol 6: 41.
Garcia-Crespo D, Juste RA, Hurtado A, 2005. Selection of ovine housekeeping genes for normalisation by real-time RT-PCR; analysis of PrP gene expression and genetic susceptibility to scrapie. BMC Vet Res 1: 3.
Garrels JI, Gibson W, 1976. Identification and characterization of multiple forms of actin. Cell 9: 793–805.
Glare EM, Divjak M, Bailey MJ, Walters EH, 2002. beta-Actin andGAPDH housekeeping gene expression in asthmatic airways is variable and not suitable for normalising mRNA levels. Thorax 57: 765–770.
Goossens K, Van Poucke M, Van Soom A, Vandesompele J, Van Zeveren A, Peelman LJ, 2005. Selection of reference genes for quantitative real-time PCR in bovine preimplantation embryos. BMC Dev Biol 5: 27.
Hamalainen HK, Tubman JC, Vikman S, Kyrola T, Ylikoski E, Warrington JA, Lahesmaa R, 2001. Identification and validation of endogenous reference genes for expression profiling of T helper cell differentiation by quantitative real-time RT-PCR. Anal Biochem 299: 63–70.
Holland NT, Smith MT, Eskenazi B, Bastaki M, 2003. Biological sample collection and processing for molecular epidemiological studies. Mutat Res 543: 217–234.
Hocquette JF Lehnert S, Barendse W, Cassar-Malek I, Picard B, 2007. Recent advances in cattle functional genomics applied to beef quality. Animal 1: 159–173.
Janovick-Guretzky NA, Dann HM, Carlson DB, Murphy MR, Loor JJ, Drackley JK, 2007. Housekeeping gene expression in bovine liver is affected by physiological state, feed intake, and dietary treatment. J Dairy Sci 90: 2246–2252.
Livak KJ, Schmittgen TD, 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408.
Nygard AB, Jorgensen CB, Cirera S, Fredholm M, 2007. Selection of reference genes for gene expression studies in pig tissues using SYBR green qPCR. BMC Mol Biol 8: 67.
Perez-Novo CA, Claeys C, Speleman F, Van Cauwenberge P, Bachert C, Vandesompele J, 2005. Impact of RNA quality on reference gene expression stability. Biotechniques 39: 52, 54, 56.
Pfaffl MW, 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45.
Pfaffl MW, Horgan GW, Dempfle L, 2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30: e36.
Radonic A, Thulke S, Mackay IM, Landt O, Siegert W, Nitsche A, 2004. Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun 313: 856–862.
Rhoads RP, McManaman C, Ingvartsen KL, Boisclair YR, 2003. The housekeeping genesGAPDH and cyclophilin are regulated by metabolic state in the liver of dairy cows. J Dairy Sci 86: 3423–3429.
Robinson TL, Sutherland IA, Sutherland J, 2007. Validation of candidate bovine reference genes for use with real-time PCR. Vet Immunol Immunopathol 115: 160–165.
Svobodová K, Bilek K, Knoll A, 2008. Verification of reference genes for relative quantification of gene expression by real-time reverse transcription PCR in the pig. J Appl Genet 49: 263–265.
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F, 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lisowski, P., Pierzchała, M., Gościk, J. et al. Evaluation of reference genes for studies of gene expression in the bovine liver, kidney, pituitary, and thyroid. J Appl Genet 49, 367–372 (2008). https://doi.org/10.1007/BF03195635
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF03195635