Abstract
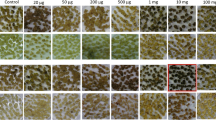
We have used two aminoglycosides, G418 and paromomycin, to develop a reliable selection system fornptll transgenic sweet-potato (Ipomoea batatas (L.) Lam.). Embryogenic calli derived from shoot apical meristems were bombarded with gold particles coated with pCAMBIA2301, which contained thenptll andgusA genes. When compared on a kill curve that was based on calli proliferation and cell viability, G413-selection proved to be more efficient and had fewer escapes than kanamycin. These bombarded expiants were then selected on G418-containing media. The total time required from bombardment to plant establishment in soil was seven to nine months. Multiple copies of the transgene were integrated into the sweetpotato genome. Northern analysis confirmed transgene expression in the regenerated plants, and a paromomycin assay demonstrated that thenptll gene was functionally expressed in transformed sweetpotato. These molecular analyses and assays all showed that selection with G418 and paromomycin is reliable. So far, we have produced 69 transgenic events with this system, at a transformation frequency of approx. 1.1%. That efficiency is based on the number of transgenic plants obtained and the amount of calli bombarded. Thus, this selection method that combines G418 with paromomycin is now available for selectingnptll transgenic sweetpotato.
Similar content being viewed by others
Literature Cited
Cheng M, Fry JE, Pang S, Zhou H, Hironaka CM, Duncan DR, Conner TW, Wan Y (1997) Genetic transformation of wheat mediated byAgrobacterium tumefaciens. Plant Physiol115: 971–980
Chu CC, Wang CC, Sun CS, Hsu C, Yin KC, Chu CY (1975) Establishment of an efficient medium for anther culture in rice through comparative experiments on the nitrogen sources. Sci Sin18: 659–668
Dekeyser R, Claes B, Marichal M, van Montagu M, Caplan A (1989) Evaluation of selectable markers for rice transformation. Plant Physiol90: 217–223
Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA minipreparation: Version II. Plant Mol Biol Rep1: 19–21
Gama MIC, Leite JPR, Cordeiro AR, Cantliffe DJ (1996) Transgenic sweetpotato plants obtained byAgrobacterium tumefaciens-mediated transformation. Plant Cell Tiss Organ Cult43: 237–244
Gordon-Kamm WJ, Spencer TM, Mangano ML, Adams TR, Daines RJ, Start WG, O’Brien JV, Chambers SA, Adams JWR, Willets NG, Rice TB, Mackey CJ, Krueger RW, Kausch AP, Lemaux PC (1990) Transformation of maize cells and regeneration of fertile transgenic plants. Plant Cell2: 603–618
Howe A, Sato S, Dweikat I, Fromm M, Clémente T (2006) Rapid and reproducible Agrobacterium-mediated transformation of sorghum. Plant Cell Rep25: 784–791
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: ß-Glucuronidase as a versatile gene fusion marker in higher plants. EMBO J6: 3901–3907
Jones A (1980) Sweet potato,In Fehr W, Hadley HH, eds, Hybridization of Crop Plants. American Society of Agronomy, Madison, pp 645–655
Kim JH, Botella JR (2002) Callus induction and plant regeneration from broccoli (Brassiaca oleracea var. italica) for transformation. J Plant Biol45: 177–181
Kimura T, Otani M, Noda T, Ideta O, Shimada T, Saito A (2001) Absence of amylose in sweetpotato [Ipomoea batatas (L.) Lam.] following the introduction of granule-bound starch synthase I cDNA. Plant Cell Rep20: 663–666
Min SR, Jeong WJ, Lee YB, Liu JR (1998) Genetic transformation of sweetpotato by particle bombardment. Kor J Plant Tiss Cult25: 329–333
Morán R, García R, López A, ZaldúaZ, Mena J, García M, Armas R, Somonte D, Rodríguez J, Gómez M, Pimentel E (1998) Transgenic sweetpotato plants carrying the delta-endotoxin gene fromBacillus thuringiensis var.tenebhonis. Plant Sci139: 175–184
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant15: 473–497
Nehra ND, Chibbar RN, Leung N, Caswell K, Mallard C, Stein-hauer L, Baga M, Kartha KK (1994) Self-fertile transgenic wheat plants regenerated from isolated scutellar tissues following microprojectile bombardment with two distinct gene constructs. Plant J5: 285–297
Newell CA, Lowe JM, Merrywether A, Rooke LM, Hamilton WDO (1995) Transformation of sweetpotato [Ipomoea batatas (L.) Lam.] withAgrobacterium tumefaciens and regeneration of plants expressing cowpea trypsin inhibitor and snowdrop lectin. Plant Sci107: 215–227
Okada Y, Kimura T, Mori M, Nishiguchi M, Saito A, Murata T, Fukuoka H (1995) Introduction of sweetpotato feathery mottle virus severe strain (SPFMV-S) coat protein to sweetpotato and regeneration of transgenic plants by electroporation. Jpn J Breed45: 116
Okada Y, Saito A, Nishiguchi M, Kimaru T, Mori M, Hanada K, Sakai J, Miyazaki C, Matsuda Y, Murata T (2001) Virus resistance in transgenic sweetpotato [Ipomoea batatas (L.) Lam] expressing the coat protein gene of sweetpotato feathery mottle vims. Theor Appl Genet103: 743–751
Otani M, Shimada T, Kimura T, Saito A (1998) Transgenic plant production from embryogénie callus of sweetpotato [Ipomoea batatas (L.) Lam.] usingAgrobacterium tumefaciens. Plant Biotechnol15:11–16
Prakash CS, Varadarajan U (1992) Genetic transformation of sweetpotato by particle bombardment. Plant Cell Rep11: 53–57
Rajasekaran K, Hudspeth RL, Cary JW, Anderson DM, Cleveland TE (2000) High-frequency stable transformation of cotton(Gossypium hirsutum L.) by particle bombardment of embryogenie cell suspension cultures. Plant Cell Rep19: 539–545
Towill LE, Mazur P (1975) Studies on the reduction of 2,3,5-triphenyltetrazolium chloride as a viability assay for plant tissue cultures. Can J Bot53: 1097–1102
Wakita Y, Otani M, Hamada T, Mori M, Iba K, Shimada T (2001) A tobacco microsomal ω-3-fatty acid desaturase gene increases the linolenic acid content in transgenic sweetpotato [Ipomoea batatas]. Plant Cell Rep20: 244–249
Weeks JT, Anderson OD, Blechl AE (1993) Rapid production of multiple independent lines of fertile transgenic wheat(Triticum aestivum). Plant Physiol102: 1077–1084
Woolfe JA (1992) Sweetpotato, An Untapped Food Resource. Cambridge University Press, New York
Yamaguchi KI, Song GQ, Honda H (2004) EfficientAgrobacterium tumefac/ens-mediated transformation of sweetpotato [Ipomoea batatas (L.) Lam.] from stem expiants using a two-step kanamycin-hygromycin selection method. In Vitro Cell Dev Biol Plant40: 359–365
Zhang P, Puonti-Kaerlas J (2000) PIG-mediated cassava transformation using positive and negative selection. Plant Cell Rep19: 1041–1048
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shin, YM., Choe, G., Shin, B. et al. Selectionof nptll transgenic sweetpotato plants using G418 and paromomycin. J. Plant Biol. 50, 206–212 (2007). https://doi.org/10.1007/BF03030631
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF03030631