Abstract
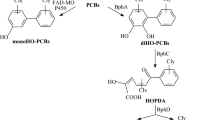
We compared the metabolism of eight di- and trichlorobiphenyls by eight bacterial strains chosen to represent a broad range of degradative activity against polychlorinated biphenyls (PCBs). The PCB congeners used were 2,3-, 2,3′-, 2,4′-, 3,3′-, 2,3,3′-, 2,4,4′-, 2,5,3′-, and 3,4,2′-chlorobiphenyl. The bacterial strains used wereCorynebacterium sp. MB1,Alcaligenes strainsA. eutrophus H850 andA. faecalis Pi434, andPseudomonas strains LB400 and H1130,P. testosteroni H430 and H336, andP. cepacia H201. The results indicated that both the relative rates of primary degradation of PCBs and the choice of the ring attacked were dependent on the bacterial strain used. The bacterial strains exhibited considerable differences in their relative reactivity preferences for attack on mono- and dichlorophenyl groups and in the degree to which the attack was affected by the chlorine substitution pattern on the nonreacting ring. For MB1 the reactivity pattern was 3-≥4-≫2-chlorophenyl with no attack on 2,4- or 2,5-chlorophenyl groups. This strain was relatively insensitive to the chlorine substitution pattern on the nonreacting ring. Strains H1130, H430, H201, and Pi434 exhibited the same reactivity preferences as MB1, but for these strains (and for all others tested) the chlorination pattern on the nonreacting ring had a strong effect. For strain H336 the reactivity preference was 4-≥2->2,4-≥3-chlorophenyl, with no evidence of attack on 2,5-chlorophenyl rings. For strains H850 and LB400 the relative reactivity was 2->2,5->3-≫2,4->4-chlorophenyl. On this basis we propose that the eight bacterial strains represent four distinct classes of biphenyl/PCB-dioxygenase activity.
The types of products formed were largely strain-independent and were determined primarily by the chlorine substitution pattern on the reacting ring. When the reacting ring was an unsubstituted phenyl or a 2-chlorophenyl group, the products were chlorobenzoic acids in high yields; for a 3-chlorophenyl ring, both chlorobenzoic acids and chloroacetophenones in moderate yields; and for a 4- or 2,4-chlorophenyl group, chlorobenzoic acids in low yields with an apparent accumulation ofmeta ring-fission product. Strains H850 and LB400 were able to degrade the 3-chlorobenzoic acid that they produced from the degradation of 2,3′-chlorobiphenyl. We conclude that despite differences among strains in the specificity of the initial dioxygenase, the specificities of the enzymes responsible for the subsequent degradation to chlorobenzoic acid and/or chloroacetophenone are quite similar for all strains.
Similar content being viewed by others
References
Ahmed, M, Focht DD (1973) Degradation of polychlorinated biphenyls by two species ofAchromobacter. Can J Microbiol 19:47–52
Barton MR, Crawford RL (1988) Novel biotransformations of 4-chlorobiphenyl by aPseudomonas sp. Appl Environ Microbiol 54:594–595
Baxter RM, Sutherland DA (1984) Biochemical and photochemical processes in the degradation of polychlorinated biphenyls Environ Sci Technol 18:608–610
Bedard DL (1990) Bacterial transformation of polychlorinated biphenyls. In: Kamely, D., Chakrabarty A, Omenn GS (eds) Biotechnology and biodegradation, Portfolio Publishing Co., The Woodlands, Texas, pp 369–388
Bedard DL, Haberl ML, May RJ, Brennan MJ (1987) Evidence for novel mechanisms of PCB metabolism inAlcaligenes eutrophus H850. Appl Environ Microbiol 53:1103–1112
Bedard DL, Unterman R, Bopp LH, Brennan MJ, Haberl ML, Johnson C (1986) Rapid assay for screening and characterizing microorganisms for the ability to degrade polychlorinated biphenyls. Appl Environ Microbiol 51:761–768
Bedard DL, Wagner RE, Brennan MJ, Haberl ML, Brown JF Jr (1987) Extensive degradation of Aroclors and environmentally transformed PCBs byAlcaligenes eutrophus H850. Appl Environ Microbiol 53:1094–1102
Bopp LH (1986) Degradation of highly chlorinated PCBs byPseudomonas strain LB400. J Ind Microbiol 1:23–29
Brown JF Jr, Bedard DL, Brennan MJ, Carnahan JC, Feng H, Wagner RE (1987) Polychlorinated biphenyl dechlorination in aquatic sediment. Science 236:709–712
Brown JF Jr, Wagner RE, Feng H, Bedard DL, Brennan MJ, Carnahan JC, May RJ (1987) Environmental dechlorination of PCBs. Environ Toxicol Chem 6:579–593
Buchanan RE, Gibbons NE (eds.) (1974) Bergey's manual of determinative bacteriology, 8th ed. The Williams & Wilkins Co, Baltimore.
Furukawa K, Hayase N, Kazunari T, Tomizuka N (1989) Molecular relationship of chromosomal genes encoding biphenyl/polychlorinated biphenyl catabolism: some soil bacteria possess a highly conservedbph operon. J Bacteriol 171:5467–5472
Furukawa K, Tomizuka N, Kamibayashi A (1979) Effect of chlorine substitution on the bacterial metabolism of various polychlorinated biphenyls. Appl Environ Microbiol 38:301–310
Furukawa K, Tonomura K, Kamibayashi A (1978) Effect of chlorine substitution on the biodegradability of polychlorinated biphenyls. Appl Environ Microbiol 35:223–227
Nadim L, Schocken MJ, Higson FK, Gibson DT, Bedard DL, Bopp LH, Mondello FJ (1987) Bacterial oxidation of polychlorinated biphenyls. In: Proceedings of the 13th Annual Research Symposium on Land Disposal, Remedial Action, Incineration, and Treatment of Hazardous Waste. U.S. Environmental Protection Agency, Cincinnati, Ohio. EPA/600/9-87/015, pp 395–402
Quensen JF III, Tiedje JM, Boyd SA (1988) Reductive dechlorination of polychlorinated biphenyls by anaerobic microorganisms from sediments. Science 242:752–754
Unterman R, Bedard DL, Brennan MJ, Bopp LH, Mondello FJ, Brooks RE, Mobley DP, McDermott JB, Schwartz CC, Dietrich DK (1988) Biological approaches for PCB degradation. In: Omenn GS et al. (eds) Reducing risks from environmental chemicals through biotechnology. Plenum Press, New York, pp 253–269
Yates JR, Mondello FJ (1989) Sequence similarities in the genes encoding polychlorinated biphenyl degradation byPseudomonas strain LB400 andAlcaligenes eutrophus H850. J Bacteriol 171:1733–1735
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Bedard, D.L., Haberl, M.L. Influence of chroline substitution pattern on the degradation of polychlorinated biphenyls by eight bacterial strains. Microb Ecol 20, 87–102 (1990). https://doi.org/10.1007/BF02543870
Issue Date:
DOI: https://doi.org/10.1007/BF02543870