Abstract
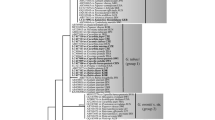
The genusMicrosphaera has been considered to be derived from sectionErysiphe of the genusErysiphe by a single event. Cleistothecial appendages are the most distinct difference between the two genera and have an important role for overwintering. To understand the phylogenetic relationship betweenErysiphe sectionErysiphe andMicrosphaera more precisely, phylogenetic trees were constructed using the nucleotide sequences of the rDNA ITS region from 11Erysiphe (sectionErysiphe) and 16Microsphaera taxa. The phylogenetic trees indicated the close relationship between the two genera. However, the generaErysiphe (sectionErysiphe) andMicrosphaera did not group into separate monophyletic lineages; instead, they formed several small clusters that were mixed together. This result suggests that the differentiations of the genera occurred two or more times independently. This also supports the idea that appendage morphology does not always accurately reflect the phylogeny of the powdery mildews because the morphology of appendages may evolve convergently under the selection pressure of their particular biotopes (host plants).
Similar content being viewed by others
Literature cited
Amano (Hirata), K. 1986. Host range and geographical distribution of the powdery mildew fungi. Japan Scientific Societies Press, Tokyo.
Blumer, S. 1933. Die Erysiphaceen Mitteleuropas unter besonderer Berücksichtigung der Schweiz. Beitr. Kryptogamenfl. Schweiz7: 1–483.
Braun, U. 1981. Taxonomic studies in the genusErysiphe I. Generic delimitation and position in the system of the Erysiphaceae. Nova Hedwig.34: 679–719.
Braun, U. 1985. Taxonomic notes on some powdery mildews (V). Mycotaxon22: 87–96.
Braun, U. 1987. A monograph of the Erysiphales (powdery mildews). Beiheft zur Nova Hedwigia89: 1–700.
Chen, G.-Q., Han, S.-J., Lai, Y.-Q., Yu, Y.-N., Zheng, R.-Y. and Zhao, Z.-Y. 1987. Flora Fungorum Sinicorum, vol. I, Erysiphales. Beijing.
Cortesi, P., Gadoury, D. M., Seem, R. C. and Pearson, R. C. 1995. Distribution and retention of cleistothecia ofUncinula necator on the bark of grapevines. Plant Dis.79: 15–19.
Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution39: 783–791.
Felsenstein, J. 1989. PHYLIP-phylogeny inference package (Version 3.2). Cladistics5: 164–166.
Gadoury, D. M. and Pearson, R. C. 1988. Initiation, development, dispersal, and survival of cleistothecia ofUncinula necator in New York vineyards. Phytopathology78: 1413–1421.
Higgins, D. G., Bleaby, A. J. and Fuchs, R. 1992. CLUSTAL V: improved software for multiple sequence alignment. Comput. Appl. Biosci.8: 189–191.
Hirata, T. and Takamatsu, S. 1996. Nucleotide sequence diversity of rDNA internal transcribed spacers extracted from conidia and cleistothecia of several powdery mildew fungi. Mycoscience37: 265–270.
Itoi, S., Nakayama, N. and Kubomura, Y. 1962. Studies on the powdery mildew disease of mulberry tree caused byPhyllactinia moricola (P. Henn.) Homma. Bull. Sericult. Exp. Stn.17: 321–445 (In Japanese with English summary.)
Katumoto, K. 1973. Notes on the generaLanomyces Gäum. andCystotheca Berk. et Curt. Rep. Tottori Mycol. Inst.10: 437–446.
Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol.16: 111–120.
Kishino, H. and Hasegawa, H. 1989. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in Hominoidea. J. Mol. Evol.29: 170–179.
Kusaba, M. and Tsuge, T. 1995. Phylogeny of Alternaria fungi known to produce host-specific toxins on the basis of variation in internal transcribed spacers of ribosomal DNA. Curr. Genet.28: 491–498.
Maddison, W. P. and Maddison, D. R. 1992. MacClade: analysis of phylogeny and character evolution. Sinauer Associates, Inc., Sunderland, MA.
Nomura, Y. 1997. Taxonomical study of Erysiphaceae of Japan. Yokendo, Tokyo (In Japanese.)
Pearson, R. C. and Gadoury, D. M. 1987. Cleistothecia, the source of primary inoculum for grape powdery mildew in New York. Phytopathology77: 1509–1514.
Saitou, N. and Nei, M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol.4: 406–425.
Sato, Y. 1990. Morphological characters of conidia and conidial germ tubes of the powdery mildew fungi on leguminous plants in Japan. Trans. Mycol. Soc. Japan31: 287–300.
Swofford, D. L. 1993. Phylogenetic Analysis Using Parsimony (PAUP, version 3.1.1). Illinois Natural History Survey, Champaign, Illinois.
Takamatsu S., Hirata, T. and Sato, Y. 1998. Phylogenetic analysis and predicted secondary structures of the rDNA internal transcribed spacers of the powdery mildew fungi (Erysiphaceae). Mycoscience39: 99–111.
Walsh, P. S., Metzger, D. A. and Higuchi, R. 1991. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Bio Techniques10: 506–513.
White, T. J., Bruns, T. D., Lee, S. and Taylor, J. 1990. Amplification and direct sequencing of fungal ribosomal genes for phylogenetics. In: PCR Protocols, (ed. by Innis, M. A., Gelfand, D. H., Sninsky, J. J. and White, T. J.), pp. 315–322. Academic Press, San Diego, California.
Zheng, R.-Y. 1983. Revision ofErysiphe glycines Tai. Mycotaxon18: 139–144.
Author information
Authors and Affiliations
Corresponding author
Additional information
Contribution No. 144 from the Laboratory of Plant Pathology, Mie University.
About this article
Cite this article
Takamatsu, S., Hirata, T., Sato, Y. et al. Phylogenetic relationships ofMicrosphaera andErysiphe sectionErysiphe (powdery mildews) inferred from the rDNA ITS sequences. Mycoscience 40, 259–268 (1999). https://doi.org/10.1007/BF02463963
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF02463963