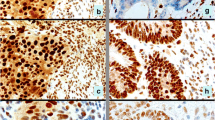
Abstract
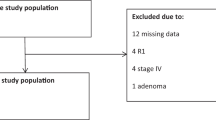
PURPOSE: The Bethesda guidelines were developed for selection of patients whose tumors should be tested for high microsatellite instability. This study examined the validity of the different Bethesda criteria in relation to microsatellite instability status to simplify their use in clinical practice. METHODS: A total of 164 patients with colorectal or hereditary nonpolyposis colorectal cancer-associated cancers were registered on the basis of the Amsterdam criteria without age limitations (11 cases), multiple tumors (2 cases), the accumulation of colorectal cancer in the family (no first-degree relatives affected or the index patient's age up to 50 years; 45 cases), an early age at onset up to 50 years (13 cases), morphologic and histopathologic manifestations (right-sided colorectal cancer, mucinous undifferentiated histology; 1 case), and the Bethesda criteria (92 cases). The microsatellite instability status of tumors was determined using the International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer marker reference panel. RESULTS: When applying all Bethesda criteria, high microsatellite instability tumors were identified in our hereditary nonpolyposis colorectal cancer registry with a sensitivity of 87 percent. Twenty-nine percent (27/92) of the Bethesda-positive patients displayed high microsatellite instability compared with 6 percent of patients (4/72) not meeting these criteria (P<0.001). Only Bethesda Criteria 1, 3, and 4 showed a significantly different distribution of the microsatellite instability status when compared with those of the remaining patients registered (P≤0.001). These three criteria detected high microsatellite instability tumors in 48 percent (10/21), 50 percent (18/36), and 31 percent (21/67) of patients, respectively. When applying these criteria only, a cumulative detection rate of 77 percent of all (24/31) high microsatellite instability cases was found, thereby identifying 89 percent of high microsatellite instability tumors among the Bethesda-positive patients. Patients matching Criteria 1, 3, and 4 frequently showed hMSH2 or hMLH1 germline mutations and tumor-specific loss of protein expression. CONCLUSION: In our hereditary nonpolyposis colorectal cancer registry the complete Bethesda criteria showed the highest sensitivity to identify patients with high microsatellite instability tumors. However, for general medical practice outside academic centers, three criteria are reasonably accurate for adequate high microsatellite instability tumor selection.
Similar content being viewed by others
References
Olschwang S. Germline mutation and genome instability. Eur J Cancer Prev 1999;9(Suppl 1):S33–7.
Fishel R, Lescoe MK, Rao MR,et al. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell 1993;75:1027–38.
Bronner CE, Baker SM, Morrison PT,et al. Mutation in the DNA mismatch-repair homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature 1994;368:258–61.
Nicolaides NC, Papadopoulos N, Liu B,et al. Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature 1994;371:75–80.
Miyaki M, Konishi M, Tanaka K,et al. Germline mutation of MSH6 as the cause of hereditary nonpolyposis colorectal cancer. Nat Genet 1997;17:271–2.
Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science 1993;260:816–9.
Lothe RA, Peltomaki P, Meling GI,et al. Genomic instability in colorectal cancer: relationship to clinicopathological variables and family history. Cancer Res 1993;53:5849–52.
Aaltonen LA, Peltomaki P, Mecklin JP,et al. Replication errors in benign and malignant tumors from hereditary nonpolyposis colorectal cancer patients. Cancer Res 1994;54:1645–8.
Lu SL, Kawabata M, Imamura T,et al. HNPCC associated with germline mutation in the TGF-beta type II receptor gene. Nat Genet 1998;19:17–8.
Rodriguez-Bigas MA, Boland CR, Hamilton SR,et al. A National Cancer Institute workshop on hereditary nonpolyposis colorectal cancer syndrome: meeting highlights and Bethesda guidelines. J Natl Cancer Inst 1997;89:1758–62.
Brown SR, Finan PJ, Cawkwell L, Quirke P, Bishop DT. Frequency of replication errors in colorectal cancer and their association with family history. Gut 1998;43:553–7.
Aaltonen LA, Salovaara R, Kristo P,et al. Incidence of hereditary nonpolyposis colorectal cancer and feasibility of molecular screening for the disease. N Engl J Med 1998;338:1481–7.
Lamberti C, Kruse R, Ruelfs C,et al. Microsatellite instability—a useful diagnostic tool to select patients at high risk for hereditary non-polyposis colorectal cancer: a study in different groups of patients with colorectal cancer. Gut 1999;44:839–43.
Vasen HF, Mecklin JP, Khan PM, Lynch HT. The International Collaborative Group on Hereditary Nonpolyposis Colorectal Cancer (ICG-HNPCC). Dis Colon Rectum 1991;34:424–5.
Watson P, Lynch HT. Extracolonic cancer in hereditary nonpolyposis colorectal cancer. Cancer 1993;71:677–85.
Watson P, Lynch HT. The tumor spectrum in HNPCC. Anticancer Res 1994;14:1635–9.
Vasen HF, Watson P, Mecklin JP, Lynch HT. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative Group on HNPCC. Gastroenterology 1999;116:1453–6.
Cravo ML, Fidalgo PO, Lage PA,et al. Validation and simplification of Bethesda guidelines for identifying apparently sporadic forms of colorectal carcinoma with microsatellite instability. Cancer 1999;85:779–85.
Messerini L, Vitelli F, De Vitis LR,et al. Microsatellite instability in sporadic mucinous colorectal carcinomas: relationship to clinico-pathological variables. J Pathol 1997;182:380–4.
Boland CR, Thibodeau SN, Hamilton SR,et al. A National Cancer Institute workshop on microsatellite instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 1998;58:5248–57.
Sutter C, Gebert J, Bischoff P,et al. Molecular screening of potential HNPCC patients using a multiplex PCR system. Mol Cell Probes 1999;13:157–65.
Kolodner RD, Hall NR, Lipford J,et al. Structure of the human MLH1 locus and analysis of a large hereditary nonpolyposis colorectal carcinoma kindred for mlh1 mutations. Cancer Res 1995;55:242–8.
Kolodner RD, Hall NR, Lipford J,et al. Structure of the human MSH2 locus and analysis of two Muir-Torre kindreds for msh2 mutations. Genomics 1994;24:516–26.
Agresti A. Categorical data analysis. New York: John Wiley & Sons, 1990.
Coughlin SS, Trock B, Criqui MH,et al. The logistic modeling of sensitivity, specificity, and predictive value of a diagnostic test. J Clin Epidemiol 1991;45:1–7.
Farrington SM, Lin-Goerke J, Ling J,et al. Systematic analysis of hMSH2 and hMLH1 in young colon cancer patients and controls. Am J Hum Genet 1998;63:749–59.
Liu B, Parsons R, Papadopoulos N,et al. Analysis of mismatch-repair genes in hereditary non-polyposis colorectal cancer patients. Nat Med 1996;2:169–74.
Nystrom-Lahti M, Wu Y, Moisio AL,et al. DNA mismatch-repair gene mutations in 55 kindreds with verified or putative hereditary non-polyposis colorectal cancer. Hum Mol Genet 1996;5:763–9.
Möslein G, Tester DJ, Lindor NM,et al. Microsatellite instability and mutation analysis of hMSH2 and hMLH1 in patients with sporadic, familial and hereditary colorectal cancer. Hum Mol Genet 1996;5:1245–52.
Wijnen JT, Vasen HF, Khan PM,et al. Clinical findings with implications for genetic testing in families with clustering of colorectal cancer. N Engl J Med 1998;339:511–8.
Wijnen J, Khan PM, Vasen H,et al. Hereditary nonpolyposis colorectal cancer families not complying with the Amsterdam criteria show extremely low frequency of mismatch-repair-gene mutation. Am J Hum Genet 1997;61:329–35.
Loukola A, de la Chapelle A, Aaltonen LA. Strategies for screening for hereditary nonpolyposis colorectal cancer. J Med Genet 1999;36:819–22.
Liu B, Farrington SM, Petersen GM,et al. Genetic instability occurs in the majority of young patients with colorectal cancer. Nat Med 1995;1:348–52.
Lynch HT, Smyrk TC, Watson P,et al. Genetics, natural history, tumor spectrum and pathology of hereditary nonpolyposis colorectal cancer: an updated review. Gastroenterology 1993;104:1535–49.
Pedroni M, Tamassia MG, Percesepe A,et al. Microsatellite instability in multiple colorectal cancers. Int J Cancer 1999;81:1–5.
Sengupta SB, Yiu CY, Boulos PB,et al. Genetic instability in patients with metachronous colorectal cancers. Br J Surg 1997;84:996–1000.
Masubuchi S, Konishi F, Togashi K,et al. The significance of microsatellite instability in predicting the development of metachronous multiple colorectal carcinomas in patients with nonfamilial colorectal carcinoma. Cancer 1999;85:1917–24.
Peel DJ, Ziogas A, Fox EA,et al. Characterization of hereditary nonpolyposis colorectal cancer families from a population-based series of cases. J Natl Cancer Inst 2000;92:1517–22.
Lynch ED, Ostermeyer EA, Lee MK,et al. Inherited mutations in PTEN that are associated with breast cancer, cowden disease, and juvenile polyposis. Am J Hum Genet 1997;61:1254–60.
Woodford-Richens K, Bevan S, Churchman M,et al. Analysis of genetic and phenotypic heterogeneity in juvenile polyposis. Gut 2000;46:656–60.
Author information
Authors and Affiliations
Additional information
Supported by the Deutsche Krebshilfe, Grant 70-1940-GF I.
About this article
Cite this article
Wüllenweber, HP., Sutter, C., Autschbach, F. et al. Evaluation of bethesda guidelines in relation to microsatellite instability. Dis Colon Rectum 44, 1281–1289 (2001). https://doi.org/10.1007/BF02234785
Issue Date:
DOI: https://doi.org/10.1007/BF02234785