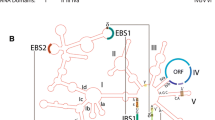
Abstract
The Zygnematales (Charophyta) contain a group-I intron (subgroup ICl) within their nuclear-encoded small subunit ribosomal DNA (SSU rDNA) coding region. This intron, which is inserted after position 1506 (relative to the SSU rDNA ofEscherichia coli), is proposed to have been vertically inherited since the origin of the Zygnematales approximately 350–400 million years ago. Primary and secondary structure analyses were carried out to model group-I intron evolution in the Zygnematales. Secondary structure analyses support genetic data regarding sequence conservation within regions known to be functionally important for in vitro self-splicing of group-I introns. Comparisons of zygnematalean group-I intron secondary structures also provided some new insights into sequences that may have important roles in in vivo RNA splicing. Sequence analyses showed that sequence divergence rates and the nucleotide compositions of introns and coding regions within any one taxon varied widely, suggesting that the “1506” group-I introns and rDNA coding regions in the Zygnematales evolve independently.
Similar content being viewed by others
References
Bhattacharya D, Surek B, Rtising M, Damberger S, Melkonian M (1994) Group-I introns are inherited through common ancestry in the nuclear-encoded rRNA of Zygnematales (Charophyceae). Proc Natl Acad Sci USA 91:9916–9920
Burke JM (1988) Molecular genetics of group-I introns: RNA structures and protein factors required for splicing — a review. Gene 73:273–294
Burke JM, Belfort M, Cech TR, Davies RW, Schweyen RJ, Shub DA, Szostak JW, Tabak HF (1987) Structural conventions for group-I introns. Nucleic Acids Res 15:7217–7221
Cech TR (1988) Conserved sequences and structures of group-I introns: building an active site for RNA catalysis — a review. Gene 73:259–271
Cech TR (1990) Self-splicing of group-I introns. Annu Rev Biochem 59:543–568
Cech TR, Damberger SH, Gutell RR (1994) Representation of the secondary and tertiary structure of group-I introns. Nature Struc Biol 1:273–280
Collins RA (1988) Evidence of natural selection to maintain a functional domain outside of the “core” in a large subclass of group-I introns. Nucleic Acids Res 16:2705–2715
Davies RW, Waring RB, Ray JA, Brown TA, Scazzocchio C (1982) Making ends meet: a model for RNA splicing in fungal mitochondria. Nature 300:719–724
De Jonckheere JF, Brown S (1994) Loss of the ORF in the SSU rDNA group-I intron of oneNaegleria lineage. Nucleic Acids Res 22:3925–3927
Dujon B (1989) Group-I introns as mobile genetic elements: facts and mechanistic speculations — a review. Gene 82:91–114
Hall DH, Povinelli CM, Ehrenman K, Pedersen-Lane J, Chu F, Belfort M (1987) Two domains for splicing in the intron of the phage T4td gene established by non-directed mutagenesis. Cell 48:63–71
Jukes TH, Cantor CR (1969) Evolution of protein molecules. In: Munoro HN (ed) Manual of protein metabolism. Academic Press, New York
Kimura M, Ohta T (1972) On the stochastic model for estimation of mutational distance between homologous proteins. J Mol Evol 2:87–90
Lambowitz AM, Belfort M (1993) Introns as mobile genetic elements. Annu Rev Biochem 62:587–622
Li W-H, Graur D (1991) Fundamentals of molecular evolution. Sinauer, Sunderland
Li W-H, Tanimura M (1987) The molecular clock runs more slowly in man than in apes and monkeys. Nature 326:93–96
Michel F, Cummings DJ (1985) Analysis of class-I introns in a mitochondrial plasmid associated with senescence ofPodospora anserina reveals extraordinary resemblance to theTetrahymena ribosomal intron. Curr Genet 10:69–79
Michel F, Westhof E (1990) Modelling the three-dimensional architecture of group-I catalytic introns based on comparative sequence analysis. J Mol Biol 216:585–610
Michel F, Hanna M, Green R, Bartel DP, Szostak JW (1989) The guanosine binding site of theTetrahymena ribozyme. Nature 342:391–395
Mix M (1980) Liste der Sammlung von Conjugaten-Kulturen im Institut für Allgemeine Botanik der Universität Hamburg Mitt Staatsinst. Allg Bot Hamburg 14:135–169
Murphy FL, Cech TR (1994) GAAA tetraloop and conserved bulge stabilize tertiary structure of a group-I intron domain. J Mol Biol 236:49–63
Oliveira MC, Ragan MA (1994) Variant forms of a self-splicing group-I intron in nuclear small-subunit rRNA genes of the marine red algaPorphyra spiralis var.amplifolia. Mol Biol Evol 11:195–207
Ragan MA, Bird CJ, Rice EL, Singh RK (1993) The nuclear 18s ribosomal RNA gene of the red algaHildenbrandia rubra contains a group-I intron. Nucleic Acids Res 211:3898
Saldanha R, Mohr G, Belfort M, Lambowitz AM (1993) Group-I and group-II introns. FASEB J 7:15–24
Sarich VM, Wilson AC (1973) Generation time and genomic evolution in primates. Science 179:1144–1147
Sogin ML, Edman JC (1989) A self-splicing intron in the small subunit rRNA gene ofPneumocystis carinii. Nucleic Acids Res 17:5349–5359
Surek B, Beemelmanns U, Melkonian M, Bhattacharya D (1994) Ribosomal RNA sequence comparisons demonstrate an evolutionary relationship between Zygnematales and Charophytes. Syst Evol 191:171–181
Vader A, Naess J, Haugli K, Haugli F, Johansen S (1994) Nuclear introns fromPhysarum polycephalum contain insertion elements that may explain how mobile group-I introns gained their open reading frames. Nucleic Acids Res 22:4553–4559
Wang Y-H, Murphy FL, Cech TR, Griffith JD (1994) Visualization of a tertiary structural domain of theTetrahymena group-I intron by electron microscopy. J Mol Biol 236:64–71
Yamada T, Tamura K, Aimi T, Songsri P (1994) Self-splicing group-I introns in eukaryotic viruses. Nucleic Acids Res 22:2532–2537
Author information
Authors and Affiliations
Additional information
Communicated by R. W. Lee
Rights and permissions
About this article
Cite this article
Bhattacharya, D., Damberger, S., Surek, B. et al. Primary and secondary structure analyses of the rDNA group-I introns of the Zygnematales (Charophyta). Curr Genet 29, 282–286 (1996). https://doi.org/10.1007/BF02221559
Received:
Issue Date:
DOI: https://doi.org/10.1007/BF02221559