Abstract
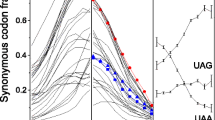
It is shown that synonymous codon usage is less biased in favor of those codons preferred by highly expressed genes at the end ofEscherichia coli genes than in the middle. This appears to be due to the close proximity of manyE. coli genes. It is shown that a substantial number of genes overlap either the Shine-Dalgarno sequence or the coding sequence of the next gene on the chromosome and that the codons that overlap have lower synonymous codon bias than those which do not. It is also shown that there is an increase in the frequency of A-ending codons, and a decrease in the frequency of G-ending codons at the end ofE. coli genes that lie close to another gene. It is suggested that these trends in composition could be associated with selection against the formation of mRNA secondary structure near the start of the next gene on the chromosome. Stop codon use is also affected by the close proximity of genes; many genes are forced to use TGA and TAG stop codons because they terminate either within the Shine-Dalgarno or coding sequence of the next gene on the chromosome. The implications these results have for the evolution of synonymous codon use are discussed.
Similar content being viewed by others
References
Arkov AL, Korolev SV, Kisselev LL (1993) Termination of translation in bacteria may be modulated via specific interaction between peptide chain release factor 2 and the last peptidyl-tRNASer/Phe. Nucleic Acids Res 21:2891–2897
Brown CM, Stockwell PA, Trotman CNA, Tate WP (1990) The signal for the termination of protein synthesis in procaryotes. Nucleic Acids Res 18:2079–2086
Bulmer M (1988) Are codon usage patterns in unicellular organisms determined by a mutation-selection balance? J Evol Biol 1:15–26
Bulmer M (1991) The selection-mutation-drift theory of synonymous codon usage. Genetics 129:897–907
de Smit MH, van Druin J (1990) Secondary structure of the ribosome binding site determines translational efficiency: a quantitative analysis. Proc Natl Acad Sci USA 87:7668–7672
Eyre-Walker A, Bulmer M (1993) Reduced synonymous substitution rate at the start of enterobacterial genes. Nucleic Acids Res 21:4599–4603
Eyre-Walker A (1995) The distance betweenEscherichia coli genes is related to gene expression levels. J Bacteriol 177:5368–5369
Freier SM, Kierzek R, Jaeger JA, Sugimoto N, Caruthers MH, Neilson T, Turner DH (1986) Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci USA 83:9373–9377
Guttman DS, Dykhuizen DE (1994) Detecting selective sweeps in naturally occurringEscherichia coli. Genetics 138:993–1003
Gold L (1988) Posttranscriptional regulatory mechanisms inEscherichia coli. Ann Rev Biochem 57:199–233
Grosjean H, Fiers W (1982) Preferential codon usage in prokaryotic genes: the optimal codon-anti-codon interaction energy and the selective codon usage in efficiently expressed genes. Gene 18:199–209
Hall BG, Sharp PM (1991) Molecular population genetics ofEscherichia coli: DNA sequence diversity at thecelC, crr, andgutB loci of natural isolates. Mol Biol Evol 9:654–665
Ikemura T (1985) Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol 2:13–34
Jaeger JA, Turner DH, Zuker M (1989) Improved prediction of secondary structures for RNA. Proc Natl Acad Sci USA 86:7706–7710
Konigsberg WJN, Godson GN (1983) Evidence for the use of rare codons in thednaG gene and other regulatory genes ofEscherichia coli. Proc Natl Acad Sci USA 80:687–691
Kurland CG (1991) Codon bias and gene expression. FEBS Lett 285:165–169
Lawrence JG, Hartl DL (1991) Unusual codon bias occurring within insertion sequences inEscherichia coli. Genetica 84:23–29
Rudd KE (1992) Alignment ofE. coli DNA sequences to a revised integrated genomic restriction map. In: Miller J (ed) A short course in bacterial genetics: a laboratory manual and handbook forEscherichia coli and related bacteria. Cold Spring Harbor Press, Cold Spring Harbor
Schurr T, Nadir E, Margalit H (1993) Identification and characterization ofE. coli ribosomal binding sites by free energy computation. Nucleic Acids Res 21:4019–4023
Sharp PM (1989) Evolution at silent-sites in DNA. In: Hill WG, McKay TFC (eds) Evolution and animal breeding: reviews of molecular and quantitative approaches in honour of Alan Robertson. CAB International, Wallingford, p 23
Sharp PM, Li W-H (1987) The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acid Res 15:1281–1295
Sharp PM, Bulmer M (1988) Selective differences among translation termination codons. Gene 63:141–145
Shields DC (1990) Switches in species-specific codon preferences: the influence of mutation biases. J Mol Evol 31:71–80
Shine J, Dalgarno L (1974) The 3′-terminal sequence ofEscherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci USA 71:1342–1346
Vallanoweth RL, Rabinowitz JC (1992) The influence of ribosome-binding-site elements on the translational efficiency inBacillus subtilis andEscherichia coli in vivo. Mol Microbiol 6:1105–1114
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Eyre-Walker, A. The close proximity ofEscherichia coli genes: Consequences for stop codon and synonymous codon use. J Mol Evol 42, 73–78 (1996). https://doi.org/10.1007/BF02198830
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF02198830