Summary
The three-dimensional structure of goose-type lysozyme (GEWL), determined by x-ray crystallography and refined at high resolution, has similarities to the structures of hen (chicken) eggwhite lysozyme (HEWL) and bacteriophage T4 lysozyme (T4L). The nature of the structural correspondence suggests that all three classes of lysozyme diverged from a common evolutionary precursor, even though their amino acid sequences appear to be unrelated (Grütter et al. 1983).
In this paper we make detailed comparisons of goose-type, chicken-type, and phage-type lysozymes. The lysozymes have undergone conformational changes at both the blobal and the local level. As in the globins, there are corresponding α-helices that have rigid-body displacements relative to each other, but in some cases corresponding helices have increased or decreased in length, and in other cases there are helices in one structure that have no counterpart in another.
Independent of the overall structural correspondence among the three lysozyme backbones is another, distinct correspondence between a set of three consecutive α-helices in GEWL and three consecutive α-helices in T4L. This structural correspondence could be due, in part, to a common energetically favorable contact between the first and the third helices.
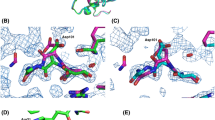
There are similarities in the active sites of the three lysozymes, but also one striking difference. Glu 73 (GEWL) spatially corresponds to Glu 35 (HEWL) and to Glu 11 (T4L). On the other hand, there are two aspartates in the GEWL active site, Asp 86 and Asp 97, neither of which corresponds exactly to Asp 52 (HEWL) or Asp 20 (T4L). (The discrepancy in the location of the carboxyl groups is about 10 Å for Asp 86 and 4 Å for Asp 97.) This lack of structural correspondence may reflect some differences in the mechanisms of action of three lysozymes. When the amino acid sequences of the three lysozyme types are aligned according to their structural correspondence, there is still no apparent relationship between the sequences except for possible weak matching in the vicinity of the active sites.
Similar content being viewed by others
References
Anderson WF, Grütter MG, Remington SJ, Matthews BW (1981) Crystallographic determination of the mode of binding of oligosaccharides to T4 bacteriophage lysozyme: implications for the mechanism of catalysis. J Mol Biol 147:523–543
Argos P, Rossmann MG (1979) Structural comparisons of hemebinding proteins. Biochemistry 18:4951–4960
Argos P, Tsukihara T, Rossmann MG (1980) A structural comparison of concanavalin A and tomato bushy stunt virus. J Mol Evol 15:169–179
Arnheim N, Inouye M, Law L, Laudin A (1973) Chemical studies on the enzymatic specificity of goose egg white lysozyme. J Biol Chem. 248:233–236
Artymiuk PJ, Blake CCF, Sippel AE (1981) Genes pieced together-exons delineate homologous structures of diverged lysozymes. Nature 290:287–288
Banner DW, Bloomer AC, Petsko GA, Phillips DC, Pogson CI, Wilson IA, Corran PH, Furth AI, Milman JD, Offord RE, Priddle J, Waley SC (1975) Structure of chicken muscle triose phosphate isomerase determined crystallographically at 2.5 Å resolution. Nature 255:609–614
Bernard O, Hozumi N, Tonegawa S (1978) Sequences of mouse immunoglobulin light chain genes before and after somatic changes. Cell 15:1133–1144
Blake CCF (1978) Do genes-in-pieces imply proteins-in-pieces? Nature 273:267
Blake CCF, Koenig DF, Mair GA, North ACT, Phillips DC, Sarma VR (1965) Structure of hen egg-white lysozyme. A three-dimensional Fourier synthesis at 2 Å resolution. Nature 206:757–761
Canfield RE, McMurry S (1967) Purification and characterization of a lysozyme from goose egg-white. Biochem Biophys Res Commun 26:38–42
Canfield RD, Kammesman S, Sobel JH, Morgan FJ (1971) Primary structure of lysozymes from man and goose. Nature (New Biol) 232:16–17
Chothia C, Levitt M, Richardson D (1977) Structure of proteins: packing of α-helices and pleated sheets. Proc Natl Acad Sci USA 74:4130–4134
Craik CS, Buchman SR, Beychok S (1980) Intron-exon splice junctions map at protein surfaces. Proc Natl Acad Sci USA 77:1384–1388
Craik CS, Buchman SR, Beychok S (1981) O2 binding properties of the product of the central exon of β-globin gene. Nature 291:87–90
Craik CS, Sprang S, Fletterick R, Rutter WJ (1982) Intronexon splice junctions map at protein surfaces. Nature 299:180–182
Fitch WM (1966) An improved method of testing for evolutionary homology. J Mol Biol 16:9–16
Ford LO, Johnson LN, Machin PA, Phillips DC, Tjian R (1974) Crystal structure of a lysozyme-tetrasaccharide lactone complex. J Mol Biol 88:349–371
Gilbert W (1978) Why genes in pieces? Nature 271:501
Gö M (1983) Molecular structural units, exons, and function of chicken lysozyme. Proc Natl Acad Sci USA 80:1964–1968
Grütter MG, Matthews BW (1982) Amino acid substitutions far from the active site of bacteriophage T4 lysozyme reduce catalytic activity and suggest that the C-terminal lobe of the enzyme participates in substrate binding. J Mol Biol 154:525–535
Grütter MG, Weaver LH, Matthews BW (1983) Goose lysozyme structure: an evolutionary link between hen and bacteriophage lysozymes? Nature 303:828–831
Harada H, Sarma R, Kakudo M, Hara S, Ikenaka T (1981) The three-dimensional structure of the lysozyme produced byStreptomyces erythraeus. J Biol Chem 256:11600–11602
Imoto I, Johnson LN, North ACT, Phillips DC, Rupley J (1972) Vertebrate lysozymes. In: Boyer P (ed) The enzymes, vol. 7, 3rd ed. Academic Press, New York, pp 665–868
Inouye M, Tsugita A (1966) The amino acid sequence of T4 bacteriophage lysozyme. J Mol Biol 22:193–196
Isaacs NW, Machin KJ, Masakuni M (1985) The three-dimensional structure of the goose-type lysozyme from the egg-white of the black swan,Cygnus atratus. Australian J Biol Sci, in press
Jollès J, Schoentgen F, Jollès P (1981) Les lysozymes de type different ont-ils un precurseur commun? C R Hebd Seances Acad Sci (Paris) 292:891–892
Jollès P, Schoentgen F, Jollès J, Dobson DE, Prager EM, Wilson AC (1984) Stomach lysozymes of ruminants II> Amino acid sequence of cow lysozyme 2 and immunological comparisons with other lysozymes. J Biol Chem 259:11617–11625
Jung A, Sippel AE, Grez M, Schultz G (1980) Exons encode functional and structural units of chicken lysozyme. Proc Natl Acad Sci USA 77:5759–5763
Kleppe G, Vasstrand E, Jensen HB (1981) The specificity requirements of bacteriophage T4 lysozyme. Involvement ofN-acetamido groups. Eur J Biochem 119:589–593
Lebioda L, Hatada M, Tulinsky A, Mavridis IM (1982) comparison of the folding of 2-keto-3-deoxy-6-phosphogluconate aldolase, triosephosphate isomerase and pyruvate kinase. Implications in molecular evolution. J Mol Biol 162:445–458
Lesk AM, Chothia C (1980) How different amino acid sequences determine similar protein structures: the structure and evolutionary dynamics of the globins. J Mol Biol 136:225–270
Levine M, Muirhead H, Stammers DK, Stuart DI (1978) Structure of pyruvate kinase and similarities with other enzymes: possible implications for protein taxonomy and evolution. Nature 271:626–630
Liljas L, Unge T, Fridborg K, Jones TA, Lovgren S, Skoglund U, Strandberg B (1982) Structure of satellite tobacco necrosis virus at 3.0Å resolution. J Mol Biol 159:93–108
Matthews BW (1977) X-ray structure of proteins. In: Neurath H, Hill RL (eds) The proteins, vol 3. 3rd ed. Academic Press, New York, pp 403–590
Matthews BW, Remington SJ (1974) The three-dimensional structure of the lysozyme from bacteriophage T4. Proc Natl Acad Sci USA 71:4178–4182
Matthews BW, Rossmann MG (1985) Comparison of protein structures. Methods Enzymol, in press
Matthews BW, Grütter MG, Anderson WF, Remington SJ (1981a) Common precursor of lysozymes from hen egg-white and bacteriophage T4. Nature 290:334–335
Matthews BW, Remington SJ, Grütter MG, Anderson WF (1981b) Relation btween hen egg-white lysozyme and bacteriophage T4 lysozyme: evolutional implications. J Mol Biol 147:545–558
McLachlan AD (1979) Gene duplications in the structural evolution of chymotrypsin. J Mol Biol 128:49–79
Ohlendorf DH, Anderson WF, Lewis M, Pabo CO, Matthews BW (1983) Comparsion of the structures of cro and “ repressor proteins from bacteriophage “. J Mol Biol 164:757–769
Ploegman JH, Drent G, Kalk KH, Hol WGJ (1978) Structure of bovineliver rhodanese. I. Structure determination at 2.5 Å resolution and a comparison of the conformation and sequence of its two domains. J Mol Biol 123:557–594
Quinto C, Quiroga M, Swain WF, Nikovits WC Jr, Standring DN, Pictet R, Valenzuela P, Rutter WJ (1982) Rat preprocarboxypeptidase A: cDNA sequence and preliminary characterization of the gene. Proc Natl Acad Sci USA 79:31–35
Rao ST, Rossmann MG (1973) Comparison of super-secondary structuresin proteins. J Mol Biol 76:241–256
Remington SJ, Matthews BW (1978) A general method to assess the similarity of protein structures with applications to T4 bacteriophage. Proc Natl Acad Sci USA 75:2180–2184
Remington SJ, Matthews BW (1980) A systematic approach to the comparison of protein structures. J Mol Biol 140:77–99
Remington SJ, Ten Eyck LF, Matthews BW (1977) Atomic coordinates for T4 phage lysozyme. Biochem Biophys Res Commun 75:265–269
Remington SJ, Anderson WF, Owen J, Ten Eyck LF, Grainger CT, Matthews BW (1978) The structure of the lysozyme from bacteriophage T4: an electron density map at 2.4 Å resolution. J Mol Biol 118:81–91
Richardson JS, Richardson DC, Thomas KA, Silverton EW, Davies DR (1976) Similarity of three-dimensional structure between the immunoglobulin domain and the copper, zinc superoxide dismutase subunit. J Mol Biol 102:221–235
Rossmann MG (1979) Processing oscillation diffraction data for very large unit cells with an automatic convolution technique and profile fitting. J Appl Crystallogr 12:225–238
Rossmann MG, Argos P (1976) A comparison of the heme binding pocket in globins and cytochrome b5. J Biol Chem 250:7525–7532
Rossmann MG, Argos P (1976) Exploring structural homology of proteins. J Mol Biol 105:75–96
Rossmann MG, Argos P (1977) The taxonomy of protein structure. J Mol Biol 109:99–129
Rossmann MG, Moras D, Olsen KW (1974) Chemical and biological evolution of a nucleotide-binding protein. Nature 250:194–199
Rossmann MG, Abad-Zapatero C, Murthy MRN, Liljas, L, Jones TA, Strandberg B (1983) Structural comparisons of some small spherical plant viruses. J Mol Biol 165:711–736
Sarma R, Harada S, Tanaka N, Kakudo M, Hara S, Ikenaka T (1979) Structure ofStreptomyces erythraeus lysozyme at 6 Å resolution. J Biochem (Tokyo) 86:1765–1771
Schmid MF, Weaver LH, Holmes MA, Grütter MG, Ohlendorf DH, Reynolds RA, Remington SJ, Matthews BW (1981) An oscillation data collection system for high-resolution protein crystallography. Acta Crystallogr A37:701–710
Schoentgen F, Jollés J, Jollés P (1982) Complete amino acid sequence of ostrich (Struthio camelus) egg-white lysozyme, a goose-type lysozyme. Eur J Biochem 123:489–497
Sharon N, Eshdat Y, Maoz I, Bernstein Y, Prager EM, Wilson AC (1974) Comparative studies of the active site region of lysozymes from eleven different sources. Isr J Chem 12:591–603
Simpson RJ, Morgan FJ (1983) Complete amino acid sequence of Embden Goose (Anser anser) egg-white lysozyme. Biochim Biophys Acta 744:349–351
Simpson RJ, Begg GS, Dorow DS, Morgan FJ (1980) Complete amino acid sequence of the goose-type lysozyme from the egg white of the black swan. Biochemistry 19:1814–1819
Stein IP, Catterall JF, Kristo P, Means AR, O'Malley BW (1980) Ovomucoid intervening sequences specify functional domains and generate protein polymorphism. Cell 21:681–687
Steitz TA, Ohlendorf DH, McKay DB, Anderson WF, Matthews BW (1982) Structural similarity in the DNA-binding domains of catabolite gene activator andcro repressor proteins. Proc Natl Acad Sci USA 79:3097–3100
Tang JJN, James MNG, Hsu IN, Jenkins JA, Blundell TA (1978) Structural evidence for gene duplication in the evolution of the acid proteases. Nature 271:618–621
Wierenga RK, Drenth J, Schultz GE (1983) Comparison of the three-dimensional protein and nucleotide structure of the FAD-binding domain ofp-hydroxybenzoate hydroxylase with the FAD- as well as NADPH-binding domains of glutathione reductase. J Mol Biol 167:725–739
Yaguchi M, Roy C, Rollin CF, Paice MG, Jurasek L (1983) A fungal cellulase shows sequence homology with the active site of hen egg-white lysozyme. Biochem Biophys Res Commun 116:408–411
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Weaver, L.H., Grütter, M.G., Remington, S.J. et al. Comparison of goose-type, chicken-type, and phage-type lysozymes illustrates the changes that occur in both amino acid sequence and three-dimensional structure during evolution. J Mol Evol 21, 97–111 (1985). https://doi.org/10.1007/BF02100084
Received:
Revised:
Issue Date:
DOI: https://doi.org/10.1007/BF02100084