Abstract
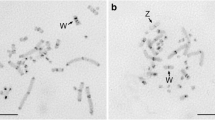
We compared the organization of satellite DNA (stDNA) and its chromosomal allocation inMus domesticus and inMus musculus. The two stDNAs show similar restriction fragment profiles after digestion (probed withM. domesticus stDNA) with some endonucleases of which restriction sequences are present in the 230–240 bp repetitive unit of theM. domesticus stDNA. In contrast, EcoRI digestion reveals thatM. musculus stDNA lacks most of the GAATTC restriction sites, particularly at the level of the half-monomer. The chromosome distribution of stDNA (revealed by anM. domesticus stDNA probe) shows different patterns in theM. domesticus andM. musculus karyotypes, with about 60% ofM. domesticus stDNA retained in theM. musculus genome. It is particularly noteworthy that the pericentromeric regions ofM. musculus chromosomes 1 and X are totally devoid ofM. domesticus stDNA sequences. In both groups, the differences in energy transfer between the stDNA-bound fluorochromes Hoechst 33258 and propidium iodide suggest that AT-rich repeated sequences have a much more clustered array in theM. domesticus stDNA, as if they are organized in tandem repeats longer than those ofM. musculus. Considering the data as a whole, it seems likely that the evolutionary paths of the two stDNAs diverged after the generation of the ancestral 230–240 bp stDNA repetitive unit through the amplification, in theM. domesticus genome, of a family repeat which included the EcoRI GAATTC restriction sequence.
Similar content being viewed by others
References
Bonhomme F, Catalan J, Britton Davidian J, Chapman VM, Moriwaki K, Nevo E, Thaler L (1984) Biochemical diversity and evolution in the genus Mus. Biochem Genet 22:275–303
Bottiroli G, Cionini PG, Docchio F, Sacchi CA (1984) In situ evaluation of the functional state of chromatin by means of Quinacrine Mustard staining and time-resolved fluorescence microscopy. Histochem J 16:223–233
Bottiroli G, Croce AC, Gerzeli G, Barni S (1989) DNA double staining for a fluorescence energy transfer study of chromatin in liver cells. Cell Biophys 15:249–263
Britton-Davidian J, Nadeau JH, Croset H, Thaler L (1989) Genic differentiation and origin of Robertsonian populations of the house mouse (Mus musculus domesticus Rutty). Genet Res 53:29–44
Brown SDM, Dover GA (1980) Conservation of segmental variants of satellite DNA of Mus musculus in a related species: Mus spretus. Nature 285:47–49
Burns J, Chan VT-W, Jonasson JA, Fleming KA, Taylor S, Mc Gee JOD (1985) Sensitive system for visualising DNA probes hybridized in situ: rapid sex determination of intact cells. J Clin Pathol 38:1085–1092
Corneo G, Ginelli E, Soave C, Bernardi G (1968) Isolation and characterization of mouse and guinea pig satellite deoxyribonucleic acid. Biochemistry 7:4373–4379
Elder FFB, Hsu TC (1988) Tandem fusion in the evolution of mammalian chromosomes. In: Daniel A (ed) The cytogenetics of mammalian autosomal rearrangements. Alan R. Liss, New York, pp 481–506
Elder FFB, Lee MR (1985) The chromosomes of Sigmodon ochrognathus and S. fulviventer suggest a realignment of Sigmodon species groups. J Mammal 66:511–518
Faircloug RH, Cantor CR (1978) The use of singlet-singlet energy transfer to study macromolecules assemblies. Methods Enzymol 48:347–379
Ferris SD, Sage RD, Prager EM, Ritte U, Wilson AC (1983) Mitochondrial DNA evolution in mice. Genetics 105:681–721
Festing MFW, Lovell DP (1981) Domestication and development of the mouse as a laboratory animal. In: Berry RJ (ed) Biology of the house mouse. Academic Press, London, New York, pp 43–62
Flam WG, McCallum M, Walker PMB (1967) The isolation of complementary strands from a mouse DNA fraction. Proc Natl Acad Sci USA 57:1729–1734
Gamperl R, Ehmann C, Bachmann K (1982) Genome size and heterochromatin variation in rodents. Genetica 58:199–212
Gropp A, Winking H (1981) Robertsonian translocations: cytology, meiosis, segregation patterns and biological consequences of heterozygosity. In: Berry RJ (ed) Biology of the house mouse. Academic Press, London, New York, pp 141–181
Hoerz W, Altenburger W (1981) The compaction of mouse heterochromatin as studied by nuclease digestion. FEBS Lett 134:25–28
Hoerz W, Zachau HG (1977) Characterization of distinct segments in mouse satellite DNA by restriction nucleases. Eur J Biochem 73:383–392
Hsu TC, Arrighi FE (1971) Distribution of constitutive heterochromatin in mammalian chromosomes. Chromosoma 34:243–253
Jones KW (1970) Chromosomal and nuclear location of mouse satellite DNA in individual cells. Nature 255:912–915
Langlois RG, Carrano AV, Gray JW, Van Dilla MA (1980) Cytochemical studies of metaphase chromosomes by flow cytometry. Chromosoma 77:229–251
Latt SA, Sahar E, Eisenhard ME (1979) Pairs of fluorescent dyes as probes of DNA and chromosomes. J Histochem Cytochem 27:65–71
Maniatis T, Fritsch EF, Sambrook J (1982) Molecular cloning. A laboratory manual. Cold Spring Harbour Laboratory, NY, p 466
Manuelidis L (1981) Consensus sequence of mouse satellite DNA indicates it is derived from base pair repeats. FEBS Lett 129:25–28
Marshall JT, Sage RD (1981) Taxonomy of the house mouse. In: Berry RJ (ed) Biology of the house mouse. Academic Press, London, New York, pp 15–25
Mayfield JE, Ellison JR (1976) The fluorescence localization of mouse satellite DNA in interphase nuclei. Chromosoma 54:27–31
Mazrimas JA, Balhorn R, Hatch FT (1979) Separation of satellite DNA chromatin and main band DNA chromatin from mouse brain. Nucleic Acids Res 7:935–946
Moriwaki K, Yonekawa H, Gotoh O, Minezawa M, Winking H, Gropp A (1984) Implications of the genetic divergence between European wild mice with Robertsonian translocations from the viewpoint of mitochondrial DNA. Genet Res 43:277–287
Nishioka Y (1987) Y-chromosomal DNA polymorphism in mouse inbred strains. Genet Res 50:69–72
Pardue ML (1985) In situ hybridization. In: Hames BD, Higgins SJ (eds) Nucleic acid hybridization. IRL Press, Oxford, pp 179–202
Pardue ML, Gall JG (1970) Chromosomal localization of mouse satellite DNA. Science 168:1356–1358
Pietras DF, Bennet KL, Siracusa LD, Woodworth-Gutai M, Chapman VM, Gross KW, Kane-Haas C, Hastie ND (1983) Construction of a small Mus musculus repetitive DNA library: identification of a new satellite sequence in Mus musculus. Nucleic Acids Res 11:6965–6983
Redi CA, Capanna E (1988) Robertsonian heterozygotes in the house mouse and the fate of their germ cells. In: Daniel A (ed) The cytogenetics of mammalian autosomal rearrangements. Alan R Liss, New York, pp 315–358
Redi CA, Garagna S, Mazzini G, Winking H (1986) Pericentromeric heterochromatin and A—T contents during Robertsonian fusion in the house mouse. Chromosoma 94:31–35
Reed Rice N, Straus NA (1973) Relatedness of mouse satellite deoxyribonucleic acid to deoxyribonucleic acid of various Mus species. Proc Natl Acad Sci USA 70:3546–3550
Schiller PW (1975) The measurement of intramolecular distances by energy transfer. In: Chen RF, Edelhoch H (eds) Biochemical fluorescence: Concepts. Marcel Dekker, New York, pp 285–303
Schmidtke J, Brennecke H, Schmid M, Neitzel H, Sperling K (1981) Evolution of muntjac DNA. Chromosoma 84:187–193
Selander RK, Yang SY (1969) Protein polymorphism and genic heterozygosity. Genetics 63:653–667
Selander RK, Hunt WG, Yang SY (1969) Protein polymorphism and genic heterozygosity in two European subspecies of the house mouse. Evolution 23:379–390
Southern EM (1975) Long range periodicities in mouse satellite DNA. J Mol Biol 94:51–69
Sparrow AH, Price HJ, Underbrink AG (1972) A survey of DNA content per cell and per chromosome of prokaryotic and eukaryotic organisms: some evolutionary considerations. In: Smith HH (ed) Evolution of genetic systems. Brookhaven symposia in biology, N. 23. Gordon and Breach, New York, pp 451–494
Sumner AT (1972) A simple technique for demonstrating centromeric heterochromatin. Exp Cell Res 75:304–306
Sutton WD, McCallum M (1972) Related satellite DNA's in the genus Mus. J Mol Biol 71:633–656
Thaler L, Bonhomme F, Britton Davidian J (1981) Processes of speciation and semi-speciation in the house mouse. In: Berry RJ (ed) Biology of the house mouse. Academic Press, New York, pp 27–39
Wahl GM, Stern M, Stark GR (1979) Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl paper and rapid hybridization by using dextransulphate. Proc Natl Acad Sci USA 76:3683–3687
Waring M, Britten RJ (1966) Nucleotide sequence repetition: a rapidly reassociating fraction of mouse DNA. Science 154:791–794
Wigler M, Sweet R, Sim GK, Wold B, Pellicer A, Lacy E, Maniatis T, Silvestein S, Axel R (1979) Transformation of mammalian cells with genes from procaryotes and eucaryotes. Cell 16:777–785
Yonekawa H, Moriwaki K, Gotoh O, Miyashita N, Migita S, Bonhomme F, Hjorth JP, Petras ML, Tagashira Y (1982) Origins of laboratory mice deduced from restriction patterns of mitochondrial DNA. Differentiation 22:222–226
Yunis G, Yasmineh WG (1971) Heterochromatin, satellite DNA and cell function. Science 174:1200–1209
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Redi, C.A., Garagna, S., Della Valle, G. et al. Differences in the organization and chromosomal allocation of satellite DNA between the European long tailed house miceMus domesticus andMus musculus . Chromosoma 99, 11–17 (1990). https://doi.org/10.1007/BF01737284
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF01737284