Summary
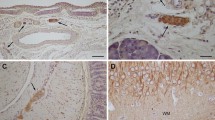
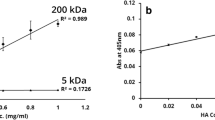
Members of the CD44 family of cell surface hyaluronate-binding proteins have been implicated in cell migration, cell-matrix interactions and tumor progression. To determine whether these proteins might play a role in the normal functions of Schwann cells and in their tumorigenesis, we examined the patterns of CD44 expression in Schwann cells from rat peripheral nerve, rat Schwann cell tumor lines, and human schwannomas. Normal rat spinal nerves and primary Schwann cell cultures expressed standard CD44 (CD44s) but not alternatively spliced variant isoforms. In contrast, rat Schwann cell tumor lines expressed both CD44s and a number of variants, including proteins containing sequences encoded by exon v6. Furthermore, we found that these cell lines bind hyaluronate, and that their cell surface hyaluronate binding correlates with CD44 expression. All of the human schwannomas also expressed CD44 variants, especially epitopes encoded by exon v5, the border between v7 and v8, and v9-10. These data indicate that Schwann cells normally express CD44s, that Schwann cell tumors express both CD44s and particular variants of CD44, and that CD44s and possibly variants of CD44 are involved in hyaluronate recognition by Schwann cell tumors.
Similar content being viewed by others
References
Le Douarin NM, Dulac C, Dupin E, Cameron-Curry P: Glial cell lineages in the neural crest. Glia 4:175–184, 1991
Jessen KR, Mirsky R: Schwann cells: Early lineage, regulation of proliferation and control of myelin formation. Curr Opin Neurobiol 2: 575–581, 1992
Heumann R, Korsching S, Bandtlow C, Thoenen H: Changes of nerve growth factor synthesis in nonneuronal cells in response to sciatic nerve transsection. J Cell Biol 104: 1623–1631, 1987
Russel DS, Rubinstein LJ: Pathology of tumors of the nervous system. 5th ed. Baltimore Williams & Wilkins Co. 1989, pp. 533–571
Jacoby LB, MacCollin M, Louis DN, Mohney T, Rubio M, Pulaski K, Trofatter JA, Kley N, Seizinger B, Ramesh V, Gusella JF: Exon scanning for mutation of the NF2 gene in schwannomas. Hum Mol Gen 3: 413–419, 1994
Irving RM, Moffat DA, Hardy DG, Barton DE, Xuereb JH, Maher ER: Somatic NF2 gene mutations in familial and non-famial vestibular schwannoma. Hum Mol Gen 3: 347–350, 1994
Twist EC, Ruttledge MH, Rousseau M, Sanson M, Papi L, Merel P, Delattre O, Thomas G, Rouleau GA: The neurofibromatosis type 2 gene is inactivated in schwannomas. Hum Mol Gen 3: 147–151, 1994
Peltonen JS, Jaakkola S, Lebwohl M, Revnall S, Riskeli L, Virkanen I, Uitto T: Cellular differentiation and expression of matrix genes in type I neurofibromatosis. Lab Invest 59: 760–771, 1988
Riccardi VM, Eichner JE: Neurofibromatosis: Phenotype, natural history and pathogenesis. Johns Hopkins Univ. Press, Baltimore, 1986
Matsui I, Tanimura M, Kobayashi N, Sawada T, Nagahara N, Akatsuka J: Neurofibromatosis type 1 and childhood cancer. Cancer 72: 2746–2754, 1993
Sasaki T, Onodera S: Glycosaminoglycans in neurofibromas. J Dermatol 17: 653–660, 1990
Kao GF, Laskin WB, Olsen TG: Solitary cutaneous plexiform neurilemmoma (schwannoma): a clinicopathologic, immunohistochemical, and ultrastructural study of 11 cases. Mod Pathol 2: 20–26, 1989
Tona A, Perides G, Rahemtulla F, Dahl D: Extracellular matrix in regenerating rat sciatic nerve: a comparative study on the localization of laminin, hyaluronic acid, and chondroitin sulfate proteoglycans, including versican. J Histochem Cytochem 41: 593–599, 1993
Picker LJ, Nakache M, Butcher EC: Monoclonal antibodies to human lymphocyte homing receptors define a novel class of adhesion molecules in diverse cell types. J Cell Biol 109: 927–937, 1989
Vogel H, Butcher EC, Picker LJ: H-CAM expression in the human nervous system: evidence for a role in diverse glial interactions. J Neurocytol 21: 363–373, 1992
Stamenkovic I, Aruffo A, Amiot M, Seed B: The hematopoietic and epithelial forms of CD44 are distinct polypeptides with different adhesion potentials for hyaluronatebearing cells. EMBO J 10: 343–348, 1991
Hofmann M, Rudy W, Zöller M, Tölg C, Ponta H, Herrlich P, Günthert U: CD44 splice variants confer metastatic behavior in rats: homologous sequences are expressed in human tumor cell lines. Cancer Res 51: 5292–5297, 1991
Dougherty GJ, Lansdorp PM, Cooper DL, Humphries RK: Molecular cloning of CD44R1 and CD44R2, two novel isoforms of the human CD44 lymphocyte ‘homing” receptor expressed by hemopoietic cells. J Exp Med 174: 1–5, 1991
Screaton GR, Bell MV, Jackson DG, Cornelis FB, Gerth U, Bell JI: Genomic structure of DNA encoding the lymphocyte homing receptor CD44 reveals at least 12 alternatively spliced exons. Proc Natl Acad Sci USA 89:12160–12164, 1992
Tölg C, Hofmann M, Herrlich P, Ponta H: Splicing choice from ten variant exons establishes CD44 variability. Nucleic Acids Res 21: 1225–1229, 1993
Heider K-H, Hofmann M, Horst E, van den Berg F, Ponta H, Herrlich P, Pals ST: A human homologue of the rat metastasis-associated variant of CD44 is expressed in colorectal carcinomas and adenomatous polyps. J Cell Biol 120: 227–233, 1993
Sherman L, Sleeman J, Herrlich P, Ponta H: Hyaluronate receptors: key players in growth, differentiation, migration and tumor progression. Curr Opin Cell Biol 6: 726–733, 1994
Lesley J, Hyman R, Kincade PW: CD44 and its interaction with the extracellular matrix. Adv Immunol 54: 271–335, 1993
Bartolazzi A, Peach R, Aruffo A, Stamenkovic I: Interaction between CD44 and hyaluronate is directly implicated in the regulation of tumor development. J Exp Med 180: 53–66, 1994
Günthert U, Hofmann M, Rudy W, Reber S, Zöller M, Haűmann I, Matzku S, Wenzel A, Ponta H, Herrlich P: A new variant of glycoprotein CD44 confers metastatic potential to rat carcinoma cells. Cell 65: 13–24, 1991
Rudy W, Hofmann M, Schwartz-Albiez R, Zöller M, Heider K-H, Ponta H, Herrlich P: The two major CD44 proteins expressed on a metastatic rat tumor cell line are derived from different splice variants: each one individually suffices to confer metastatic behavior. Cancer Res 53: 1262–1268, 1993
Ponta H, Hofmann M, Herrlich P: Recent advances in the genetics of metastasis. Eur J Cancer (in press), 1994
Wielenga VJM, Heider K-H, Offerhaus GJA, Adolf GR, van den Berg FM, Ponta H, Herrlich P, Pals ST: Expression of CD44 variant proteins in human colorectal cancer is related to tumor progression. Cancer Res 53: 4754–4756, 1993
Sinn H-P, Heider K-H, Skroch-Angel P, von Minckwitz G, Kaufmann M, Herrlich P, Ponta H: Human mammary carcinomas express homologues of metastasis-associated variants of CD44. Breast Cancer Res Treat (in press), 1994
Paterson DJ, Jefferies WA, Green JR, Brandon MR, Corthesy P, Puklavec M, Williams AF: Antigens of activated rat T lymphocytes including a molecule of 50,000 Mr detected only on CD4 positive T blasts. Mol Immunol 24: 1281–1290, 1987
Matzku S, Wenzel A, Liu S, Zöller M: Antigenic differences between metastatic and nonmetastatic BSp73 rat tumor variants characterized by monoclonal antibodies. Cancer Res 49: 1294–1299, 1989
Koopman G, Heider K-H, Horst E, Adolf GR, van den Berg F, Ponta H, Herrlich P, Pals ST: Activated human lymphocytes and aggressive non-Hodgkin lymphomas express a homologue of the rat metastasis-associated variant of CD44. J Exp Med 177: 897–904, 1993
Guerci A, Monge M, Baron-Van Evercooren A, Lubetzki C, Dancea S, Boutry JM, Goujet-Zalc C, Zalc B: Schwann cell marker defined by a monoclonal antibody (224-58) with species cross-reactivity. I. Cellular localization. J Neurochem 46: 425–434, 1986
Allen RW, Trach KA, Hoch JA: Identification of the 37-kDa protein displaying a variable interaction with the erythroid cell membrane as glyceraldehyde-3-phosphate dehydrogenase. J Biol Chem 262: 649–653, 1987
Zhou DFH, Ding JF, Picker LJ, Bargatze RF, Butcher EC, Goeddel DV: Molecular cloning and expression of Pgp-1: The mouse homolog of the human H-CAM (Hermes) lymphocyte homing receptor. J Immunol 143: 3390–3395, 1989
Schubert D, Heinemann S, Carlisle W, Tarikas H, Kimes B, Patrick J, Steinbach JH, Culp W, Brandt BL: Clonal cell lines from the rat central nervous system. Nature 249: 224–227, 1974
Pfeiffer SE, Wechsler W: Biochemically differentiated neoplastic clone of Schwann cells. Proc Natl Acad Sci USA 69: 2885–2889, 1972
Tomozawa Y, Sueoka N:In vitro segregation of different cell lines with neuronal and glial properties from a stem cell line of rat neurotumor RT4. Proc Natl Acad Sci USA 75: 6305–6309, 1978
Langford LA, Porter S, Bunge RP: Immortalized rat Schwann cells produce tumorsin vivo. J Neurocytol 17:521–529, 1988
Turley EA: Proteoglycans and cell adhesion. Their putative role during tumorigenesis. Cancer Metastasis Rev 3: 325–339, 1984
Bertrand P, Girard N, Delpech B, Duval C, D'Anjou J, Dauce JP: Hyaluronan (hyaluronic acid) and hyaluronectin in the extracellular matrix of human breast carcinomas: comparison between invasive and non-invasive areas. Int J Cancer 52: 1–6, 1992
Toole BP, Biswas C, Gross J: Hyaluronate and invasiveness of the rabbit V2 carcinoma. Proc Natl Acad Sci USA 76: 6299–6303
Knudsen CB, Knudson W: Similar epithelial-stromal interactions in the regulation of hyaluronate production during limb morphogenesis and tumor invasion. Cancer Lett 52: 113–122
He Q, Lesley J, Hyman R, Ishihara K, Kincade PW: Molecular isoforms of murine CD44 and evidence that the membrane proximal domain is not critical for hyaluronate recognition. J Cell Biol 119: 1711–1719, 1992
Hua Q, Knudson CB, Knudson W: Internalization of hyaluronan by chondrocytes occurs via receptor-mediated endocytosis. J Cell Sci 106: 365–375, 1993
Toyama-Sorimachi N, Miyasaka M: A novel ligand for CD44 is sulfated proteoglycan. Int Immunol 6: 655–660, 1994
Culty M, Nguyen HA, Underhill CB: The hyaluronan receptor (CD44) participates in the uptake and degradation of hyaluronan. J Cell Biol 116: 1055–1062, 1992
Jalkanen S, Jalkanen M: Lymphocyte CD44 binds the COOH-terminal heparin-binding domain of fibronectin. J Cell Biol 116: 817–825, 1992
Ishii S, Ford R, Thomas P, Nachman A, Steele G Jr, Jessup JM: CD44 participates in the adhesion of human colorectal carcinoma cells to laminin and type IV collagen. Surg Oncol 2: 255–264, 1993
Bargmann CI, Hung MC, Weinberg RA: Multiple independent activations of the neu oncogene by a point mutation altering the transmembrane domain of p185. Cell 45: 649–657, 1986
Perantoni AO, Rice JM, Reed CD, Watatani M, Wenk ML: Activatedneu oncogene sequences in primary tumors of the peripheral nervous system induced in rats by transplacental exposure to ethylnitrosourea. Proc Natl Acad Sci USA 84: 6317–6321, 1987
Nikitin AY, Ballering LAP, Lyons J, Rajewsky MF: Early mutation of theneu (erbB-2) gene during ethylnitrosourea-induced oncogenesis in the rat Schwann cell lineage. Proc Natl Acad Sci USA 88: 9939–9943, 1991
Dougall WC, Qian X, Peterson NC, Miller MJ, Samanta A, Greene MI: The neu-oncogene: signal transduction pathways, transformation mechanisms and evolving therapies. Oncogene 9: 2109–2123, 1994
Bargmann CI, Weinberg RA: Increased tyrosine kinase activity with the protein encoded by the activatedneu oncogene. Proc Natl Acad Sci USA 85: 5394–5398, 1988
Nakamura T, Ushijima T, Ishizaka Y, Nagao M, Nemoto T, Hara M, Ishikawa T:neu proto-oncogene mutation is specific for the neurofibromas in aN-nitroso-N-ethylurea-induced hamster neurofibromatosis model but not for hamster melanomas and human Schwann cell tumors. Cancer Res 54: 876–980, 1994
Trofatter JA, MacCollin MM, Rutter JL, Murrell JR, Duyao MP, Parry DM, Eldridge R, Kley N, Menon AG, Pulaski K, Haase VH, Ambrose CM, Munroe D, Bove C, Haines JL, Martuza RL, MacDonald ME, Seizinger BR, Short MP, Buckler AJ, Gusella JF: A novel moesin-, ezrin-, radixinlike gene is a candidate for the neurofibromatosis 2 tumor suppressor. Cell 72: 791–800, 1993
Tsukita S, Oishi K, Sato N, Sagara J, Kawai A, Tsukita S: ERM family members as molecular linkers between the cell surface glycoprotein CD44 and actin-based cytoskeletons. J Cell Biol 126: 391–401, 1994
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Sherman, L., Skroch-Angel, P., Moll, J. et al. Schwann cell tumors express characteristic patterns of CD44 splice variants. J Neuro-Oncol 26, 171–184 (1995). https://doi.org/10.1007/BF01052620
Issue Date:
DOI: https://doi.org/10.1007/BF01052620