Abstract
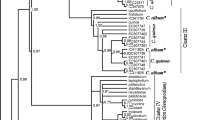
Sequences of the nrDNA internal transcribed spacer (ITS) region and the cpDNAtrnL/trnF intergenic spacer (IGS) region were analysed for 41 Mediterranean and Eurasian representatives of the Anthemideae (Compositae) to ascertain the systematic position of the unispecific genusCastrilanthemum Vogt & Oberprieler and to study the phylogeny of the Anthemideae in the Mediterranean region. Both markers consistently point to the close relationship ofCastrilanthemum with the W Mediterranean generaLeucanthemopsis (Giroux) Heywood andProlongoa Boiss., forming a strongly supported monophyletic group (theLeucanthemopsis-group which also comprises the unispecific genusHymenostemma (Kunze) Willk.). Results also demonstrate that subtribes Achilleinae, Leucantheminae, and Matricariinae sensu Bremer & Humphries are non-monophyletic. Besides results from sequence variation, a marked 5bp-deletion intrnL/trnF IGS divides all these subtribes into more basal subgroups which are related to each other and to the large Eurasian generaTanacetum L. andAnthemis L., and a monophyletic group of closely related and more advanced subgroups which also contain the monophyletic Chrysantheminae sensu Bremer & Humphries. For this second group a W Mediterranean centre of diversification is suggested, however, its sister-group relationships within the basal grade of generic groups remain unclear.
Similar content being viewed by others
References
Bayer R. J., Starr J. R. (1998) Tribal phylogeny of the Asteraceae based on two non-coding chloroplast sequences, thetrnL intron andtrnL/trnF intergenic spacer. Ann. Missouri Bot. Gard. 85: 242–256.
Bremer K. (1988) The limits of amino acid sequence data in angiosperm phylogenetic reconstruction. Evolution 42: 795–803.
Bremer K. (1994) Asteraceae, Cladistics & classification. Portland.
Bremer K., Humphries Ch. J. (1993) Generic monograph of the Asteraceae-Anthemideae. Bull. Brit. Mus. (Nat. Hist.) Bot. 23: 71–177.
Eriksson T., Wikström N. (1995) AutoDecay ver. 3.0. — Stockholm: Computer program distributed by the authors, Botaniska Institutionen, Stockholm University.
Facius D., Fartmann B., Huber J., Nikoleit K., Schondelmaier J., Steinhauser S. (1999) Sequencing brochure — Instructions for DNA template preperation, primer design, and sequencing with the LI-COR DNA sequencer 4000 and 4200 series. Version 4. — MWG-Biotech AG, Ebersberg.
Felsenstein J. (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791.
Francisco-Ortega J., Jansen R. K., Crawford D. J., Santos-Guerra A. (1995) Chloroplast DNA evidence for intergeneric relationships of the Macaronesian endemic genusArgyranthemum (Asteraceae). Syst. Bot. 20: 413–422.
Francisco-Ortega J., Santos-Guerra A., Hines A., Jansen R. K. (1997) Molecular evidence for a Mediterranean origin of the Macaronesian endemic genusArgyranthemum (Asteraceae). Amer. J. Bot. 84: 1595–1613.
Fujii N., Ueda K., Watano Y., Shimizu T. (1997) Intraspecific sequence variation of chloroplast DNA inPedicularis chamissonis Steven (Scrophulariaceae) and geographic structuring of the Japanese “alpine” plants. J. Pl. Res. 110: 195–207.
Fujii N., Ueda K., Watano Y., Shimizu T. (1999) Further analysis of intraspecific sequence variation of chloroplast DNA inPrimula cuneifolia Ledeb. (Primulaceae): Implications for biogeography of the Japanese alpine flora. J. Pl. Res. 112: 87–95.
Galiano E. F., Heywood V. H. (1960) Catálogo de plantas de la provincia de Jaén (mitad oriental). Jaén.
Hellwig F. H., Nolte M., Ochsmann J., Wissemann V. (1999) Rapid isolation of total cell DNA from milligram plant tissue. Haussknechtia 7: 29–34.
Hervier J. (1905) Excursions botaniques de M. Élisée Reverchon dans le massif de La Sagra et à Vélez-Rubio (Espagne) de 1899 à 1903. Bull. Acad. Int. Géogr. Bot. 15: 1–32, 57–72, 89–120, 157–178.
Heywood V. H. (1954) A revision of the Spanish species ofTanacetum L. subsect.Leucanthemopsis Giroux. Anales Inst. Bot. Cavanilles 12: 313–374.
Heywood V. H., Humphries Ch. J. (1977) Anthemideae — systematic review. In: Heywood V. H., Harborne J. B., Turner B. L. (eds.) The biology and chemistry of the Compoitae, 2. London, New York, San Francisco, pp. 851–898.
Kim K.-J., Jansen R. K. (1994) Comparisons of phylogenetic hypothesis among different data sets in dwarf dandelions(Krigia): additional information from internal transcribed spacer sequences of nuclear DNA. Plant Syst. Evol. 190: 157–185.
Larson A. (1994) The comparison of morphological and molecular data in phylogenetic systematics. In: Schierwater B., Streit B., Wagner G. P., DeSalle R. (eds.) Molecular ecology and evolution: approaches and applications. Basel, pp. 371–390.
Mason-Gamer R. J., Kellogg E. A. (1996) Testing for phylogenetic conflict among molecular data sets in the tribe Triticeae (Gramineae). Syst. Biol. 45: 524–545.
Reitbrecht F. (1974) Fruchtanatomie und Systematik der Anthemideae (Asteraceae). Thesis (Wien).
Swofford D. L. (1993) PAUP: Phylogenetic analysis using parsimony, version 3.1.1. Illinois Natural History Survey, Champaign.
Taberlet P., Gielly L., Pautou G., Bouvet J. (1991) Universal primers for amplification of three noncoding regions of chloroplast DNA. Pl. Molec. Biol. 17: 1105–1109.
Templeton A. R. (1983) Phylogenetic inference from restriction endonuclease cleavage site maps with particular reference to the evolution of humans and the apes. Evolution 37: 221–244.
Thompson J. D., Higgins D. G., Gibson T. J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Research 22: 4673–4680.
Trehane P. (1995) Proposal to conserveChrysanthemum L. with a conserved type (Compositae). Taxon 44: 439–441.
Tutin T. G., Heywood V. H., Burges N. A., Moore D. M., Valentine D. H., Walters S. M., Webb D. A. (eds.) (1976) Flora Europaea. Vol 4. Cambridge.
Vogt R., Oberprieler Ch. (1996)Castrilanthemum Vogt & Oberprieler, a new genus of the Compositae-Anthemideae. Anales Jard. Bot. Madrid 54: 336–346.
Watson L. E., Evans T. M., Boluarte T. (2000) Molecular phylogeny and biogeography of tribe Anthemideae (Asteraceae), based on chloroplast genendhF. Mol. Phyl. Evol. 15: 59–69.
White T. J., Bruns T, Lee S., Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M., Gelfand D., Sninsky J., White T. (eds.) PCR protocols: a guide to methods and application. San Diego, pp. 315–322.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Oberprieler, C., Vogt, R. The position ofCastrilanthemum Vogt & Oberprieler and the phylogeny of Mediterranean Anthemideae (Compositae) as inferred from nrDNA ITS and cpDNAtrnL/trnF IGS sequence variation. Pl Syst Evol 225, 145–170 (2000). https://doi.org/10.1007/BF00985465
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00985465