Abstract
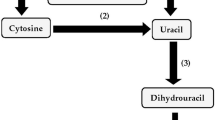
Creatinine deimination has been newly detected in the following various cytosine deaminase-forming microorganisms: Escherichia coli, Proteus mirabilis, Pseudomonas aureofaciens, Pseudomonas chlororaphis and Pseudomonas cruciviae. All these microorganisms, except for E. coli, formed cytosine deaminase in a constitutive or repressive way. P. putida 77 and E. coli showed highly increased formation of creatinine deiminase in the presence of creatinine and cytosine. Throughout serial DEAE-Sephacel and Sephacryl S-300 column chromatographies, the cytosine deaminases of these microorganisms, except for that of P. ovalis, were found to hydrolyze both creatinine and cytosine at comparable rates. No concrete evidence was obtained for the presence of any other protein that hydrolyzed creatine and/or cytosine than the cytosine deaminases in the three test microorganisms randomly selected for investigation.
Different from P. putida 77, none of the test microorganisms degraded N-methylhydantoin; neither N-methylhydantoin amidohydrolase nor N-carbamoylsarcosine amidohydrolase was formed in the presence of creatinine in these microorganisms. As a result, the wide occurrence of cytosine deaminases in microorganisms was found to be related to the wide distribution of those microorganisms which hydrolyze creatinine to N-methylhydantoin without further degradation.
Similar content being viewed by others
References
Esders TW, Lynn SY (1985) Purification and properties of creatinine iminohydrolase from Flavobacterium filamentosum. J Biol Chem 260:3915–3922
Hahn A, Schafer L (1925) The behavior of pyrimidine derivatives in organisms. II. The action of E. coli on uracil and cytosine. Z Biol 83:511–514
Ipata PL, Marmocchi F, Magni G, Felicioli R, Polidoro G (1971) Bakers's yeast cytosine deaminase. Some enzymatic properties and allosteric inhibition by nucleosides and nucleotides. Biochemistry 10:4270–4276
Kellen RA, Warren RAJ (1974) Pyrimidine metabolism in Pseudomonas acidovorans. Can J Microbiol 20:427–433
Kim JM, Shimizu S, Yamada H (1986a) Purification and characterization of a novel enzyme, N-carbamoylsarcosine amidohydrolase, from Pseudomonas putida 77. J Biol Chem 261:11832–11839
Kim JM, Shimizu S, Yamada H (1986b) Sarcosine oxidase involved in creatinine degradation in Alcaligenes denitrificans subsp. denitrificans J9, and Arthrobacter spp. J5 and J11. Agric Biol Chem 50:2811–2816
Kim JM, Shimizu S, Yamada H (1987) Evidence for the presence of a cytosine deaminase that does not catalyze the deimination of creatinine. FEBS Lett (in press)
Kun E, Kearney EB (1974) Ammonia. In: Bergmeyer HU (ed) Methods of enzymatic analysis, 2nd ed, vol 4. Verlag Chemie, Weinheim, pp 1802–1806
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–275
Mori N, Sano M, Tani Y, Yamada H (1980) Purification and properties of sarcosine oxidase from Cylindrocarpon didymum M-1. Agric Biol Chem 44:1391–1397
O'Donovan GA, Neuhard J (1970) Pyrimidine metabolism in microorganisms. Bacteriol Rev 34:278–343
Sakai T, Yu T, Taniguchi K, Omata S (1975a) Purification of cytosine deaminase from Pseudomonas aureofaciens. Agric Biol Chem 39:2015–2020
Sakai T, Yu T, Tabe H, Omata S (1975b) Purification of cytosine deaminase from Serratia marcescens. Agric Biol Chem 39:1623–1629
Shimizu S, Kim JM, Shinmen Y, Yamada H (1986) Evaluation of two alternative metabolic pathways for creatinine degradation in microorganisms. Arch Microbiol 145:329–333
Szulmajster J (1958a) Bacterial fermentation of creatinine. I. Isolation of N-methylhydantoin. J Bacteriol 75:633–639
Szulmajster J (1958b) Bacterial degradation of creatinine. II. Creatinine desimidase. Biochim Biophys Acta 30:154–163
Uwajima T, Terada O (1977) Production, purification and crystallization of creatinine deiminase of Corynebacterium lilium. Agric Biol Chem 41:330–344
West TP, O'Donovan GA (1982) Repression of cytosine deaminase by pyrimidines in Salmonella typhimurium. J Bacteriol 149:1171–1174
West TP, Shanley MS, O'Donovan GA (1982) Purification and some properties of cytosine deaminase from Salmonella typhimurium. Biochim Biophys Acta 719:251–258
Yamada H, Shimizu S, Kim JM, Shinmen Y, Sakai T (1985) A novel metabolic pathway for creatinine degradation in Pseudomonas putida putida 77. FEMS Microbiol Lett 30:337–340
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Kim, J.M., Shimizu, S. & Yamada, H. Cytosine deaminase that hydrolyzes creatinine to N-methylhydantoin in various cytosine deaminase-forming microorganisms. Arch. Microbiol. 147, 58–63 (1987). https://doi.org/10.1007/BF00492905
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00492905