Summary
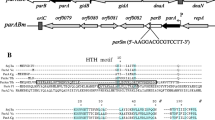
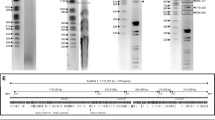
Some strains of the yeast Kluyveromyces lactis contain a pair of linear DNA plasmids, k1 and k2, 8.8 and 13.8 kilobase pairs long, respectively. Simultaneous presence of the two plasmids confer a killer phenotype on the cell by producing a toxin which blocks the growth of sensitive yeast species. Previous genetic studies have suggested that the toxin protein is coded by the k1 plasmid. We have now determined the total nucleotide sequence of k1 DNA. The genome is 8,874 base pairs in length. It contains four protein-coding reading frames, three transcribed from one strand and the fourth transcribed from the complementary strand and has terminal inverted repeats of 202 base pairs. Nuclease S1 mapping confirmed this arrangement and showed that these genes are transcribed. The terminal repeats and the four genes form an extremely compact genome, with some overlapping of genes. All four genes use highly biased codons, 86% of them having A or T at the wobble position, reminiscent of yeast mitochondrial genes. Three genes share a very similar 5′ leader sequence. The nature of gene products is discussed in the light of what is known of the excreted toxin protein.
Similar content being viewed by others
References
Anderson S, de Bruijn MHL, Coulson AR, Epron IC, Sanger F, Young IG (1982) J Mol Biol 156:687–717
Barrell BG, Air GM, Hutchinson III CA (1972) Nature 264:34–41
Berk AJ, Sharp PA (1977) Cell 12:721–732
Bevan EA, Herring AJ, Mitchell DJ (1973) Nature 245:81–86
Blobel G, Dobberstein B (1975) J Cell Biol 67:835–851
Bostian KA, Jayachandran S, Tipper DJ (1983) Cell 32:169–180
Bostian KA, Elliot Q, Bussey H, Brun V, Smith A, Tipper DJ (1984) Cell 36:741–751
Breunig KG, Dahlems U, Das S, Hollenberg CP (1984) Nucl Acids Res 12:2327–2341
Campbell A (1962) Adv Genet 11:101–145
De Louvencourt L, Fukuhara H, Heslot H, Wésolowski M (1983) J Bacteriol 154:737–742
Gunge N, Tamaru A, Ozawa F, Sakaguchi K (1981a) J Bacteriol 145:382–391
Gunge N, Sakaguchi K (1981b) J Bacteriol 147:155–160
Gunge N, Murata K, Sakaguchi K (1982) J Bacteriol 151:462–464
Gunge N (1983) Ann Rev Microbiol 37:252–276
Guthrie C, Abelson J (1982) In: Strathern JN, Jones EW, Broach JR (eds) Molecular Biology of the Yeast Saccharomyces cerevisiae. Metabolism and Gene Expression. Cold Spring Harbor Laboratory, Cold Spring Harbor New York, p 487
Hartley JL, Donelson JE (1980) Nature 286:860–865
Herring AJ, Bevan EA (1974) J Gen Virol 22:387–394
Hubbard SC, Ivatt RJ (1981) Ann Rev Biochem 50:555–583
Julius D, Blair L, Brake A, Sprague G, Thorner J (1983) Cell 32:839–852
Maxam AM, Gilbert W (1980) Methods Enzymol 65:499–558
Mizuno K, Matsuo H (1984) Nature 309:558–560
Niwa O, Sakaguchi K, Gunge N (1981) J Bacteriol 148:988–990
Perlman D, Halvorson HO (1983) J Mol Biol 167:391–409
Shinagawa M, Padamanaban R (1980) Proc Natl Acad Sci USA 77:3831–3835
Skipper N, Thomas D, Lau P (1984) EMBO journal 3:107–111
Sor F, Fukuhara H (1983a) Cell 32:391–396
Sor F, Wésolowski M, Fukuhara H (1983b) Nucleic Acids Res 11:5037–5044
Stark MJR, Mileham AJ, Romanos MA, Boyd A (1984) Nucleic Acids Res 12:6011–6030
Sugisaki Y, Gunge N, Sakaguchi K, Yamasaki K, Tamura G (1983) Nature 304:464–466
Tzagoloff A (1982) Mitochondria. Plenum Press, New York, p 308
Wésolowski M, Algeri A, Goffrini P, Fukuhara H (1982a) Current Genetics 5:191–197
Wésolowski M, Dumazert P, Fukuhara H (1982b) Curr Genet 5:199–203
Wésolowski M, Algeri A, Fukuhara H (1982c) Curr Genet 5:205–208
Wickner RB (1981) In: Strathern JN, Jones EN, Broach JR (eds) Molecular Biology of the Yeast Saccharomyces cerevisiae: Life Cycle and Inheritance. Cold Spring Harbor Laboratory, Cold Spring Harbor New York, p 415
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Sor, F., Fukuhara, H. Structure of a linear plasmid of the yeast Kluyveromyces lactis; Compact organization of the killer genome. Curr Genet 9, 147–155 (1985). https://doi.org/10.1007/BF00436963
Received:
Issue Date:
DOI: https://doi.org/10.1007/BF00436963