Summary
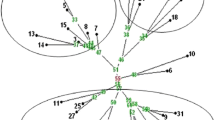
A genetic linkage map of lentil comprising 333 centimorgans (cM) was constructed from 20 restriction fragment length, 8 isozyme, and 6 morphological markers segregating in a single interspecific cross (Lens culinaris × L. orientalis). Because the genotypes at marker loci were determined for about 66 F2 plants, linkages are only reported for estimates of recombination less than 30 cM. Probes for identification of restriction fragment length polymorphisms (RFLPs) were isolated from a cDNA and EcoRI and PstI partial genomic libraries of lentil. The cDNA library gave the highest frequency of relatively low-copy-number probes. The cDNAs were about twice as efficient, relative to random genomic fragments, in RFLP detection per probe. Nine markers showed significant deviations from the expected F2 ratios and tended to show a predominance of alleles from the cultigen. Assuming a genome size of 10 Morgans, 50% of the lentil genome could be linked within 10 cM of the 34 markers and the map is of sufficient size to attempt mapping of quantitative trait loci.
Similar content being viewed by others
References
Bantle J, Maxwell I, Hahn W (1976) Specificity of oligo(dT)-cellulose chromatography in the isolation of polyadenylated RNA. Anal Biochem 72:413–427
Beckmann J, Soller M (1983) Restriction fragment length polymorphisms in genetic improvement: methodologies, mapping and costs. Theor Appl Genet 67:35–43
Bennett M, Smith J (1976) Nuclear DNA amounts in angiosperms. Proc R Soc London 274:227–274
Bernatzky R, Tanksley S (1986) Majority of random cDNA clones correspond to single loci in the tomato genome. Mol Gen Genet 203:8–14
Blixt S (1972) Mutation genetics in Pisum. Agric Hortic Genet 30:1–293
Botstein D, White R, Skolnick M, Davis R (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314–331
Chirgwin J, Przybyla A, MacDonald R, Rutter W (1979) Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry 18:5294–5299
Church G, Gilbert W (1984) Genomic sequencing. Proc Natl Acad Sci USA 81:1991–1995
Edwards M, Stuber C, Wendel J (1987) Molecular-marker-facilitated investigations of quantitative-trait loci in maize. 1. Numbers, genomic distribution, and types of gene action. Genetics 116:113–125
Feinberg A, Vogelstein B (1983) A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem 132:6–13
Figdore S, Kennard W, Song K, Slocum M, Osborn T (1988) Assessment of the degree of restriction fragment length polymorphism in Brassica. Theor Appl Genet 75:833–840
Hanahan D (1983) Studies on transformation of E. coli with plasmids. J Mol Biol 166:557–580
Helentjaris T, King G, Slocum M, Siedenstrang C, Wegman S (1985) Restriction fragment polymorphisms as probes for plant diversity and their development as tools for applied plant breeding. Plant Mol Biol 5:109–118
Helentjaris T, Slocum M, Wright S, Schaefer A, Nienhuis J (1986) Construction of genetic linkage maps in maize and tomato using restriction fragment length polymorphisms. Theor Appl Genet 72:761–769
Helentjaris T, Weber D, Wright S (1988) Identification of the genomic locations of duplicate nucleotide sequences in maize by analysis of restriction fragment length polymorphisms. Genetics 118:353–363
Ladizinsky G, Braun D, Goshen D, Muehlbauer F (1984) The biological species of the genus Lens L. Bot Gaz 145:253–261
Landry B, Michelmore R (1985) Selection of probes for restriction fragment length analysis from plant genomic clones. Plant Mol Biol Rep 3:174–179
Landry B, Kesseli R, Farrara B, Michelmore R (1987a) A genetic map of lettuce (Lactuca sativa L.) with restriction fragment length polymorphisms, isozyme, disease resistance and morphological markers. Genetics 116:331–337
Landry B, Kesseli R, Leung H, Michelmore R (1987b) Comparison of restriction endonucleases and sources of probes for their efficiency in detecting restriction fragment length polymorphisms in lettuce (Lactuca sativa L.). Theor Appl Genet 74:646–653
Lebowitz R, Soller M, Beckmann J (1987) Trait based analyses for the detection of linkage between marker loci and quantitative traits in crosses between inbred lines. Theor Appl Genet 73:556–562
Maniatis T, Fritsch E, Sambrook J (1982) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press. Cold Spring Harbor/NY
Messing J (1983) New M13 vectors for cloning. Methods Enzymol 101:20–78
Muehlbauer F, Slinkard A (1981) Genetics and breeding methodology. In: Webb C, Hawtin G (eds) Lentils. Commonwealth Agricultural Bureaux, Slough, England, pp 69–90
Muehlbauer F, Weeden N, Hoffman D (1989) Inheritance and linkage relationships of several isozyme loci in lentil (Lens Miller). J Hered (in press)
Murray M, Thompson W (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4231–4325
Myers J, Weeden N (1988) Proposed revision of guidelines for genetic analysis in Phaseolus vulgaris. Annu Rep Bean Improv Coop 31:16–19
Riggs M, McLachlan A (1986) A simplified screening procedure for large numbers of plasmid mini-preparations. Biotechniques 4:312–313
Saghai-Maroof M, Soliman K, Jorgensen R, Allard R (1984) Ribosomal DNA spacer length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci USA 81:8014–8018
Soller M, Beckmann J (1983) Genetic polymorphism in varietal identification and genetic improvement. Theor Appl Genet 67:25–33
Soltis D, Haufler C, Darrow D, Gastony G (1983) Starch gel electrophoresis of ferns: a compilation of grinding buffers, gel and electrode buffers and staining schedules. Am Fern J 73:9–27
Southern E (1975) Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol 98:503–517
Stuber C, Moll R, Goodman M, Schaffer H, Weir B (1980) Allozyme frequency changes associated with selection for increased grain yield in maize (Zea mays L.). Genetics 95:225–236
Stuber C, Goodman M, Moll R (1982) Improvement of yield and ear number resulting from selection at allozyme loci in a maize population. Crop Sci 22:737–740
Stuber C, Edwards M, Wendel J (1987) Molecular marker-facilitated investigations of quantitative trait loci in maize. 2. Factors influencing yield and its component traits. Crop Sci 27:639–648
Suiter K, Wendel J, Case J (1983) Linkage-1: a pascal computer program for the detection and analysis of genetic linkage. J Hered 74:203–204
Tanksley S, Rick C (1980) Isozymic gene linkage map of the tomato: applications in genetics and breeding. Theor Appl Genet 57:161–170
Tanksley S, Medino-Filho H, Rick C (1982) Use of naturally occurring enzyme variation to detect and map genes controlling quantitative traits in an interspecific backcross of tomato. Heredity 49:11–25
Vandenberg A (1987) Inheritance and linkage of several qualitative traits in lentil. PhD thesis, University of Saskatchewan, Saskatoon, Canada
Weeden N, Zamir D, Tadmor Y (1988) Applications of isozyme analysis in pulse crops. In: Summerfield R (ed) World crops: cool season food legumes. Nijhoff, Amsterdam, pp 979–987
Weller J, Soller M, Brody T (1988) Linkage analysis of quantitative traits in an interspecific cross of tomato (Lycopersicon esculentum × Lycopersicon pimpinellifolium) by means of genetic markers. Genetics 118:329–339
Zamir D, Ladizinsky G (1984) Genetics of allozyme variants and linkage groups in lentil. Euphytica 33:329–336
Zamir D, Tadmor Y (1986) Unequal segregation of nuclear genes in plants. Bot Gaz 147:355–358
Author information
Authors and Affiliations
Additional information
Communicated by J. Beckmann
Rights and permissions
About this article
Cite this article
Havey, M.J., Muehlbauer, F.J. Linkages between restriction fragment length, isozyme, and morphological markers in lentil. Theoret. Appl. Genetics 77, 395–401 (1989). https://doi.org/10.1007/BF00305835
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00305835