Abstract
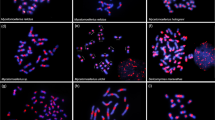
C-band variation between the Caledia taxa is extensive with numerous large interstitial and telomeric blocks of heterochromatin being present in the South-east Australian and Moreton taxa while the Torresian types possess small centromeric or telomeric C-bands. In situ hybridization using 3H-cRNA from a 168 bp (base pairs) highly repeated sequence, originally isolated from the South-east Australian taxon, defined further variation between the C. captiva taxa. This sequence family is present in each of the interstitial and telomeric constitutive heterochromatic blocks in the South-east Australian and Moreton taxa. However, it is represented in only a fraction of the heterochromatic regions, defined by C-banding, within the three Torresian types. A second, unrelated 144 bp sequence family, originally isolated from the Daintree taxon, is restricted to the procentric blocks of heterochromatin of chromosomes 2–7, 9 and 10 in the Daintree taxon. This sequence is A-T rich and possesses a region of dyad symmetry. Quantitative measurements for the two sequence families revealed a wide range of copy numbers between the C. captiva taxa. The 168 bp family has approximately 150,000, 35,000 and 4,000 copies, respectively, in the South-east Australian/ Moreton, Torresian and Daintree genomes. There are 2,000,000 and 100,000 copies of the 144 bp sequence in the Daintree and Papuan Torresian taxa, respectively. The distributional, quantitative and sequence characteristics of these repeat families imply that past amplification or introgression has played a major role in the evolution of these sequences. There is an overall negative correlation between the quantity of the 168 bp sequence and the levels of reproductive isolation and genie divergence between the various taxa. It is possible that some of the reduction in the viability of the hybrid individuals is due to the quantitative changes in these sequences. Moreover, the quantitative and qualitative characteristics of highly repeated DNA families may play a role in the modulation of such essential cellular functions as cell cycle duration, nuclear organization and gene expression.
Similar content being viewed by others
References
Andrews DL, Millstein L, Hamkalo BA, Gottesfeld JM (1984) Competition between Xenopus satellite I sequences and Pol III genes for stable transcription complex formation. Nucleic Acids Res 12:7753–7769
Appels R (1982) The molecular cytology of wheat-rye hybrids. Int Rev Cytol 80:93–132
Appels R, Peacock WJ (1978) The arrangement and evolution of highly repeated (satellite) sequences with special reference to Drosophila. Int Rev Cytol [Suppl] 8:69–126
Appels R, Driscoll C, Peacock WJ (1978) Heterochromatin and highly repeated DNA sequences in rye (Secale cereale). Chromosoma 70:67–89
Arnheim N (1983) Concerted evolution of multigene families. In: Nei M, Koehn RK (eds) Evolution of genes and proteins. Sinauer Associates, Inc., Sunderland pp 38–61
Arnold ML, Appels R, Shaw DD (1985a) The heterochromatin of grasshoppers from the Caledia captiva species complex I. Sequence evolution and conservation in a highly repeated DNA family. Mol Biol Evol (in press)
Arnold ML, Appels R, Shaw DD (1985b) Evolution and conservation in a highly repeated DNA family from the grasshopper Caledia captiva. Cytobios (in press)
Bishop JO, Robertson FW, Burns JA, Melli M (1969) Methods for the analysis of deoxyribonucleic acid — ribonucleic acid hybridization data. Biochem J 115:361–370
Bostock CJ, Christie S, Lauder IJ (1976) S-phase patterns of replication of different satellite DNAs in three species of Dipodomys (Kangaroo Rat). J Mol Biol 108:417–433
Botchan MR (1974) Bovine satellite I DNA consists of repetitive units 1,400 base pairs in length. Nature 251:288–292
Coates DJ, Shaw DD (1984) The chromosomal component of reproductive isolation in the grasshopper Caledia captiva III. Chiasma distribution patterns in a new chromosomal taxon. Heredity 53:85–100
Cohen EH, Bowman SC (1979) Detection and location of three simple sequence DNAs in polytene chromosomes from virilis group species of Drosophila. Chromosoma 73:327–355
Daly JC, Wilkinson P, Shaw DD (1981) Reproductive isolation in relation to allozymic and chromosomal differentiation in the grasshopper Caledia captiva. Evolution 35:1164–1179
Doolittle WF, Sapienza C (1980) Selfish genes, the phenotype paradigm and genome evolution. Nature 284:601–603
Dover G (1982) Molecular drive: a cohesive mode of species evolution. Nature 299:111–116
Franke WW (1974) Structure, biochemistry, and functions of the nuclear envelope. Int Rev Cytol [Suppl] 4:71–236
Gall JG, Cohen EH, Atherton DD (1974) The satellite DNAs of Drosophila virilis. Cold Spring Harbor Symp Quant Biol 38:417–421
Iida S, Meyer J, Arber W (1983) Prokaryotic IS elements. In: Shapiro JA (ed) Mobile genetic elements. Acadmic Press, New York pp 159–221
Kubinski H, Gibbs P, Kasper CB (1972) Interactions of nucleic acids and cellular membranes: behavior of membranes in CsCl density gradients. Biochim Biophys Acta 281:244–252
Lima-De-Faria A, Jaworska H (1968) Late DNA synthesis in heterochromatin. Nature 217:138–142
Messing J, Vieira J (1982) A new pair of M13 vectors for selecting either strand of double-digest restriction fragments. Gene 19:269–276
Miklos GLG, Gill AC (1981) The DNA sequences of cloned complex satellite DNAs from Hawaiian Drosophila and their bearing on satellite DNA sequence conservation. Chromosoma 82:409–427
Nagl W (1974) Role of heterochromatin in the control of cell cycle duration. Nature 249:53–54
Nei M (1972) Genetic distance between populations. Am Nat 106:283–292
Novak U (1984) Structure and properties of a highly repetitive DNA sequence in sheep. Nucleic Acids Res 12:2343–2350
Ohta T, Dover GA (1984) The cohesive population genetics of molecular drive. Genetics 108:502–521
Orgel LE, Crick FHC (1980) Selfish DNA: the ultimate parasite. Nature 284:604–607
Pardue ML, Gall JG (1970) Chromosomal localization of mouse satellite DNA. Science 168:1356–1358
Peacock WJ, Brutlag D, Goldring E, Appels R, Hinton CW, Lindsley DL (1974) The organization of highly repeated DNA sequences in Drosophila melanogaster chromosomes. Cold Spring Harbor Symp Quant Biol 38:405–421
Peacock WJ, Lohe AR, Gerlach WL, Dunsmuir P, Dennis ES, Appels R (1978) Fine structure and evolution of DNA in heterochromatin. Cold Spring Harbor Symp Quant Biol 42:1121–1135
Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74:5463–5467
Shaw DD, Coates DJ (1983) Chromosomal variation and the concept of the coadapted genome — a direct cytological assessment. In: Brandham PE, Bennett MD (eds) Kew chromosome conference II. G. Allen and Unwin, London pp 207–216
Shaw DD, Wilkinson P (1980) Chromosome differentiation, hybrid breakdown and the maintenance of a narrow hybrid zone in Caledia. Chromosoma 80:1–31
Shaw DD, Moran C, Wilkinson P (1980) Chromosomal reorganization, geographic differentiation and the mechanism of speciation in the genus Caledia: In: Blackman RL, Hewitt GM, Ashburner M (eds) Insect cytogenetics, Symp Roy Entomol Soc London, No 10. Blackwells, Oxford pp 171–194
Shaw DD, Wilkinson P, Coates DJ (1982) The chromosomal component of reproductive isolation in the grasshopper Caledia captiva II. The relative viabilities of recombinant and nonrecombinant chromosomes during embryogenesis. Chromosoma 86:533–549
Shaw DD, Coates DJ, Arnold ML, Wilkinson P (1985) Temporal variation in the chromosomal structure of a hybrid zone and its relationship to karyotypic repatterning. Heredity (in press)
Siebenlist U, Simpson RB, Gilbert W (1980) E. coli RNA polymerase interacts homologously with two different promoters. Cell 20:269–281
Smith GP (1976) Evolution of repeated DNA sequences by unequal crossover. Science 191:528–535
Southern EM (1975) Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol 98:503–517
Strachan T, Webb D, Dover G (1985) Transition stages of molecular drive in multiple-copy DNA families in Drosophila. EMBO J 4:1701–1708
Sutton WD, McCallum M (1972) Related satellite DNA's in the genus Mus. J Mol Biol 71:633–656
Trick M, Dover GA (1984) Unexpectedly slow homogenisation within a repetitive DNA family shared between two subspecies of tsetse fly. J Mol Evol 20:322–329
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Arnold, M.L., Shaw, D.D. The heterochromatin of grasshoppers from the Caledia captiva species complex. Chromosoma 93, 183–190 (1985). https://doi.org/10.1007/BF00293167
Received:
Revised:
Issue Date:
DOI: https://doi.org/10.1007/BF00293167