Abstract
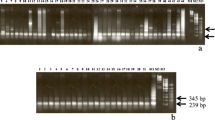
In an attempt to identify relationships among genomes of the allotetraploid Pennisetum purpureum Schumach and closely related Pennisetum species with which it can be successfully hybridized, repetitive DNA sequences were examined. Digestion with KpnI revealed two highly repetitive fragments of 140 by and 160 bp. The possibility that these sequences could be used as genome markers was investigated. Average sequences were determined for the 140 by and 160 by KpnI families from P. purpureum and P. squamulatum Fresen. Average sequences (based upon four or five repeats) were determined for the P. glaucum (L.) R. Br. 140 by KpnI family and the diploid P. hohenackeri Hochst. ex Steud. 160 bp KpnI family. The average sequences of the 160 by KpnI families in P. purpureum and P. squamulatum differ by only nine bases. The 140 by KpnI families of the three related species, P. purpureum, P. squamulantum, and P. glaucum are nearly identical, and thus likely represent a recent divergence from a common progenitor or a common genome. Each repetitive sequence may contain internal duplications, which probably diverged following amplification of the original sequence. The 140 by KpnI repeat probably evolved from the 160 by KpnI repeat since the missing 18 by segment is part of the internal duplication that is otherwise conserved in the subrepeats. Tandemly arrayed repetitive sequences in plants are likely to be composed of subrepeats which have been duplicated and amplified.
Similar content being viewed by others
References
Appels R, Moran LB, Gustafson JP (1986) Rye heterochromatin. I. Studies on clusters of the major repeating sequence and the identification of a new dispersed repetitive sequence element. Can J Genet Cytol 28:645–657
Barnes SR, James AM, Jamieson G (1985) The organization, nucleotide sequence, and chromosomal distribution of a satellite DNA from Allium cepa. Chromosoma 62:149–176
Benslimane AA, Dron M, Hartman C, Rode A (1986) Small tandemly repeated DNA sequences of higher plants likely originate from a tRNA gene ancestor. Nucleic Acids Res 14:8111–8119
Capesius I (1983) Sequence of the cryptic satellite DNA from the plant Sinapis alba. Biochem Biophys Acta 739:276–280
Dennis ES, Peacock WJ (1984) Knob heterochromatin homology in maize and its relatives. J Mol Evol 20:341–350
Dover G (1982) Molecular drive: a cohesive mode of species evolution. Nature 299:111–117
Dujardin M, Hanna WW (1983) Apomictic and sexual pearl millet × Pennisetum squamulatum hybrids. J Hered 74:277–279
Dujardin M, Hanna WW (1984) Cytogenetics of double cross hybrids between Pennisetum americanum-P. purpureum amphidiploids and P. americanum × P. purpureum × P. squamulatum interspecific hybrids. Theor Appl Genet 69:97–100
Dujardin M, Hanna WW (1989) Crossability of pearl millet with wild Pennisetum species. Crop Sci 29:77–80
Dujardin M, Hanna WW (1990) Haploid pearl millet pollen from near-tetraploid interspecific Pennisetum hybrids. Crop Sci 30:393–396
Ganal M, Hemleben V (1986) Different AT-rich satellite DNAs in Cucurbita pepo and Cucurbita maxima. Theor Appl Genet 73:129–135
Ganal M, Hemleben V (1988) Insertion and amplification of a DNA sequence in satellite DNA of Cucumis sativus L. (cucumber). Theor Appl Genet 75:357–361
Ganal M, Riede I, Hemleben V (1986) Organization and sequence analysis of two related satellite DNAs in cucumber (Cucumis sativus L.). J Mol Evol 23:23–30
Grellet F, Delcasso D, Panabieres F, Delseny M (1986) Organization and evolution of a higher plant alphoid-like satellite DNA sequence. J Mol Biol 187:495–507
Ingham LD (1990) Molecular experiments concerning the evolution of the allotetraploid Pennisetum purpureum. Dissertation, University of Florida, Gainesville, Florida
Jauhar PP (1981) Cytogenetics and breeding of pearl millet and related species. Liss, New York
Junghans H, Metzlaff M (1988) Genome specific, highly repeated sequences of Hordeum vulgare: cloning, sequencing and squash dot test. Theor Appl Genet 76:728–732
Lagudah ES, Hanna WW (1989) Species relationships in the Pennisetum gene pool: enzyme polymorphisms. Theor Appl Genet 78:801–808
Lagudah ES, Hanna WW (1990) Patterns of variation for seed proteins in the Pennisetum gene pool. J Hered 81:25–29
Leclerc RF, Siegel A (1987) Characterization of repetitive elements in several Cucurbita species. Plant Mol Biol 8:497–507
McIntyre CL, Clarke BC, Appels R (1988) Amplification and dispersion of repeated DNA sequences in the Triticeae. Plant Syst Evol 160:39–59
Maniatis T, Fritsch EF, Sambrook J (1982) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor New York
Patankar S, Joshi CP, Ranade SA, Bhave M, Ranjekar PK (1985) Interphase nuclear structure in plants: role of nuclear DNA content and highly repeated DNA sequences in chromatin structure. Proc Indian Acad Sci 94:539–551
Peacock WJ, Dennis ES, Rhoades MM, Pryor AJ (1981) Repeated DNA limited to knob heterochromatin in maize. Proc Natl Acad Sci USA 78:4490–4494
Pearson WR, Lipman DJ (1988) Improved tools for biological sequence comparison. Proc Natl Acad Sci USA 85:2444–2448
Prober JM, Trainor GL, Dam RJ, Hobbs FW, Robertson CW, Zagursky RJ, Cocuzza AJ, Jensen MA, Baumeister K (1987) A system for rapid DNA sequencing with fluorescent chainterminating dideoxynucleotides. Science 238:336–341
Rivin CT, Cullis CA, Walbot V (1986) Evaluating quantitative variation in the genome of Zea Mays. Genetics 113:1009–1019
Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74:5463–5467
Schweizer M, Ganal M, Ninnemann H, Hemleben V (1988) Species-specific DNA sequences for identification of somatic hybrids between Lycopersicon esculentum and Solanum acaule. Theor Appl Genet 75:679–684
Taylor MG, Vasil IK (1987) Analysis of DNA size, content and cell cycle in leaves of Napiergrass (Pennisetum purpureum Schumach.) Theor Appl Genet 14:681–686
Xin ZY, Appels R (1988) Occurrence of rye (Secale cereale) 350family DNA sequences in Agropyron and other Triticeae. Plant Syst Evol 160:65–76
Zhao X, Wu T, Xie Y, Wit R (1989) Genome-specific repetitive sequences in the genus Oryza. Theor Appl Genet 78:201–209
Author information
Authors and Affiliations
Additional information
Communicated by H. Saedler
Florida Aqricultural Experiment Station series #R-02758
Rights and permissions
About this article
Cite this article
Ingham, L.D., Hanna, W.W., Baier, J.W. et al. Origin of the main class of repetitive DNA within selected Pennisetum species . Molec. Gen. Genet. 238, 350–356 (1993). https://doi.org/10.1007/BF00291993
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00291993