Summary
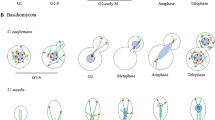
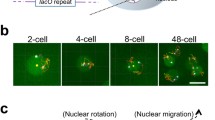
We have isolated a novel gene (NUM1) with unusual internal periodicity. The NUM1 gene encodes a 313 kDa protein with a potential Ca2+ binding site and a central domain containing 12 almost identical tandem repeats of a 64 amino acid polypeptide. num1-disrupted strains grow normally, but contain many budded cells with two nuclei in the mother cell instead of a single nucleus at the bud neck, while all unbudded cells are uninucleate: This indicates that most G2 nuclei divide in the mother before migrating to the neck, followed by the migration of one of the two daughter nuclei into the bud. Furthermore, haploid num1 strains tend to diploidize during mitosis, and homozygous num1 diploid or tetraploid cells sporulate to form many budded asci with up to eight haploid or diploid spores, respectively, indicating that meiosis starts before nuclear redistribution and cytokinesis. Our data suggest that the NUM1 protein is involved in the interaction of the G2 nucleus with the bud neck.
Similar content being viewed by others
References
Ausubel FM, Brent R, Kingston RE, Moore DO, Seidman JS, Smith JA, Struhl K (1987) Current protocols in molecular biology. John Wiley and Sons, New York
Birnboim HC, Doly J (1979) A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res 7:1513–1523
Byers B (1981) Cytology of the yeast life cycle. In: Strathern JN, Jones EW, Broach JR (eds) Molecular Biology of the Yeast Saccharomyces cerevisiae: Life cycle and Inheritance. Cold Spring Habor Laboratory Press, Cold Spring Harbor, NY, pp 59–95
Carle GF, Olson MV (1984) Separation of chromosomal DNA molecules from yeast by orthogonal-field-alternation gel electrophoresis. Nucleic Acids Res 12:5647–5664
Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinum thiocyanate-phenol-chloroform extraction. Anal Bioch 162:156–159
Frischauf AM, Lehrach H, Poustka A, Murray N (1983) Lambda replacement vectors carrying polylinker sequences. J Mol Biol 170:827–842
Hirschberg J, Simchen G (1977) Commitment to the mitotic cell cycle in yeast in relation to meiosis. Exp Cell Res 105:245–252
Hoyt MA, Stearns T, Botstein D (1988) Chromosome instability mutants of Saccharomyces cerevisiae that are defective in microtubule-mediated processes. Mol Cell Biol 10:223–234
Huffaker TC, Thomas JH, Botstein D (1988) Diverse effects of β-tubulin mutations on microtubule formation and function. J Cell Biol 106:1997–2010
Jacobs CW, Adams AEM, Szanislo PJ, Pringle JR (1988) Functions of microtubules in the Saccharomyces cerevisiae cell cycle. J Cell Biol 107:1409–1426
Kemp BE, Pearson RB (1990) Protein kinase recognition sequence motifs. Trends Biochem Sci 15:342–346
Klebe RJ, Harriss JV, Sharp ZD, Douglas MG (1983) A general method for polyethylene-glycol-induced genetic transformation of bacteria and yeast. Gene 25:333–341
Klee CB (1988) Ca2+-dependent phospholipid- (and membrane-) binding proteins. Biochemistry 27:6645–6653
Kretsinger Rh (1980) Structure and evolution of calcium-modulated proteins. CRC Crit Rev Biochem 8:119–174
Kyte J, Doolittle RF (1982) A simple method for displaying the hydrophobic character of a protein. J Mol Biol 157:105–132
Lisziewicz J, Godany A, Förster HH, Küntzel H (1987) Isolation and nucleotide sequence of a Saccharomyces cerevisiae protein kinase gene suppressing the cell cycle start mutation cdc25. J Biol Chem 262:2549–2553
Lux SE, John KM, Bennet V (1990) Analysis of cDNA for human erythrocyte ankyrin indicates a repeated structure with homology to tissue-differentiation and cell-cycle control proteins. Nature 344:36–42
Matsudaira P, Janmey P (1988) Pieces in the actin-severing protein puzzle. Cell 54:139–140
Maxam AM, Gilbert W (1980) Sequencing end-labelled DNA with base-specific chemical cleavages. Methods Enzymol 65:499–560
Nishikawa K, Ooi T (1986) Amino acid sequence homology applied to the prediction of protein secondary structure, and joint prediction with existing methods. Biochim Biophys Acta 871:45–54
Pringle JR, Hartwell LH (1981) The Saccharomyces cerevisiae cell cycle. In: Strathern JN, Jones EW, Broach JR (eds) Molecular biology of the yeast Saccharomyces cerevisiae. Life cycle and inheritance. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 97–142
Roman H, Phillips MM, Sands SM (1955) Studies of polyploid Saccharomyces. Tetraploid segregation. Genetics 40:546–561
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: A laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Schaaff I (1990) Molekulargenetische und physiologische Untersuchungen zum Pentosephosphatweg bei der Hefe Saccharomyces cerevisiae. Doctoral thesis, TH Darmstadt, FRG
Schild D, Ananthaswamy HN, Mortimer RK (1981) An endomitotic effect of a cell cycle mutation of Saccharomyces cerevisiae. Genetics 97:551–562
Schneider A, Hemphill A, Wyler T, Seebeck T (1988) Large microtubule-associated protein of T. brucei has tandemly repeated, near-identical sequences. Science 241:459–462
Sengstag C, Hinnen A (1987) The sequence of the Saccharomyces cerevisiae gene PHO2 codes for a regulatory protein with unusual aminoacid composition. Nucleic Acids Res 15:233–246
Sikorski RS, Boguski MS, Goebl M, Hieter P (1990) A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell 60:307–317
Snyder M, Davis RW (1988) SPA1: a gene important for chromosome segregation and other mitotic functions in S. cerevisiae. Cell 54:743–754
Stahl ML, Ferenz CR, Kelleher KL, Kriz RW, Knopf JL (1988) Sequence similarity of phospholipase C with the non-catalytic region of src. Nature 332:269–275
Stearns T, Hoyt MA, Botstein D (1990) Yeast mutants sensitive to antimicrotubule drugs define three genes that affect microtubule function. Genetics 124:251–262
Ursic D, Culbertson MR (1991) The yeast homology to mouse tcp-1 affects microtubule-mediated processes. Mol Cell Biol 11:2629–2640
Williamson DH, Fennell DJ (1979) Visualization of yeast mitochondrial DNA with the fluorescent stain “DAPI”. Methods Enzymol 56:728–733
Yanisch-Perron C, Vieira J, Messing J (1985) Improved M13 phage cloning vectors and host strains: nucleotide sequence of the M13mpl8 and pUC19 vectors. Gene 33:103–119
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Kormanec, J., Schaaff-Gerstenschläger, I., Zimmermann, F.K. et al. Nuclear migration in Saccbaromyces cerevisiae is controlled by the highly repetitive 313 kDa NUM1 protein. Molec. Gen. Genet. 230, 277–787 (1991). https://doi.org/10.1007/BF00290678
Received:
Issue Date:
DOI: https://doi.org/10.1007/BF00290678