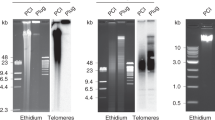
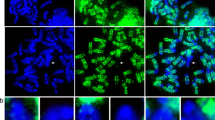
Abstract
A human interspersed repetitive DNA cloned in pBR322, the HindIII 1.9-kb (kilobase pair) sequence, was labeled with biotinylated dUTP and hybridized to acid-fixed chromosomes and paraformaldehyde-fixed whole cells in situ. Using our most sensitive detection techniques this probe highlighted on the order of 200 discrete loci, in punctate or banded arrays, that resembled a Giemsa-dark band pattern on chromosome arms. Interphase cells also displayed many discrete punctate spots of hybridization along chromosome fibers. The ubiquitous Alu sequence repeat also appeared to be concentrated in specific regions of the chromosome and predominantly highlighted Giemsa-light bands. Centromeric or ribosomal spacer DNA repeats used as controls in all studies gave the expected hybridization profiles and showed no non-specific labeling of chromosome arms. Cohesive groups of centromeric DNA arrays and rDNA clusters were observed in interphase nuclei. Refinements in methods for detecting biotin-labeled probes in situ were developed during these studies and calculations indicated that about 20 kb or more of the 1.9-kb repeat were present at each hybridization site. The chromosomal distribution of the 1.9-kb repeat suggests that this sequence may reflect, or participate in defining, ordered structureal domains along the chromosome.
Similar content being viewed by others
References
Adams J, Kaufman RE, Dretschner PJ, Harrison M, Nienhuis AW (1980) A family of long reiterated DNA sequences, one copy of which is next to the human beta globin gene. Nucleic Acids Res 8:6113–6129
Alberstson DG (1984) Localization of the ribosomal genes in Caenorhabditis elegans chromosomes by in situ hybridization using biotin-labeled probes. EMBO J 3:1227–1234
Bogenhagen DF, Sakonja S, Brown DD (1980) A control region in the center of the 5S RNA gene directs specific initiation of transcription: II. The 3′ border of the region. Cell 19:27–35
Brigati DJ, Myerson D, Leary JJ, Spalholz B, Travis SZ, Fong CKY, Hsiung GD, Ward DC (1983) Detection of viral genomes in cultured cells and paraffin embedded tissue sections using biotin-labeled hybridization probes. Virology 126:32–50
Capco DC, Krochmalnic G, Penman A (1984) A new method for preparing embeddment-free sections for transmission electron microscopy: applications to the cytoskeletal framework and other three-dimensional networks. J Cell Biol 98:1878–1885
Denhardt DT (1966) A membrane filter technique for the detection of complementary DNA. Biochim Biophys Res Comm 23:183–184
DiGiovanni L, Haynes SR, Misra R, Jelinek WR (1983) Kpn I family of long-dispersed repeated DNA sequences of man: Evidence for entry into genomic DNA of DNA copies of poly(A)-terminated Kpn I RNAs. Proc Natl Acad Sci USA 80:6533–6537
Doolittle WF, Sapienza C (1980) Selfish genes, the phenotype paradigm and genome evolution. Nature 284:601–603
Evans H, Buckland R, Pardue ML (1966) Location of the genes coding for 18S and 28S ribosomal RNA in the human genome. Chromosoma 48:405–426
Gallyas F, Gores T, Merchenthaler I (1982) High-grade intensification of the end product of the diaminobenzidine reaction for peroxidase histochemistry. J Histochem Cytochem 30:183–184
Georgiev GP, Kramerov DA, Ryskov AP, Skryabin KG, Lukanidin EM (1982) Dispersed repetitive sequences in eukaryotic genomes and their possible biological significance. Cold Spring Harbor Symp Quant Biol 47:1109–1121
Gerhard DS, Kawasaki ES, Bancroft FC, Szabo P (1981) Localization of a unique gene by direct hybridization in situ. Proc Natl Acad Sci USA 78:3755–3759
Goldman MA, Holmquist GP, Gray MC, Caston LA, Nag A (1984) Replication timing of genes and middle repetitive sequences. Science 224:686–692
Harper ME, Saunders GF (1981) Localization of single copy DNA sequences on G-banded human chromosomes by in situ hybridization. Chromosoma 83:431–439
Holmquist G, Gray M, Porter T, Jordan J (1982) Characterization of Giemsa-dark and light-band DNA. Cell 31:121–129
Inoué S (1981) Video image processing greatly enhances contrast, quality and speed in polarization-based microscopy. J Cell Biol 89:346–356
Jelinek WR, Toomey TP, Leinwand L, Duncan CH, Biro PA, Choudary PV, Weissman SM, Rubin CM, Houck CM, Deininger PL, Schmid CW (1980) Ubiquitous interspersed repeated sequences in mammalian genomes. Proc Natl Acad Sci USA 77:1398–1402
Kole LB, Haynes SR, Jelinek WR (1983) Discrete and heterogeneous high molecular weight RNAs complementary to a long dispersed repeat family (a possible transposon) of human DNA. J Mol Biol 165:257–286
Langer PR, Waldrop AA, Ward DC (1981) Enzymatic synthesis of biotin-labeled polynucleotides: novel nucleic acid affinity probes. Proc Natl Acad Sci USA 78:6633–6637
Leary JJ, Brigati DJ, Ward DC (1983) Rapid sensitive colorimetric method for visualizing biotin-labeled DNA probes hybridized to DNA or RNA immobilized on nitrocellulose: Bio-blots. Proc Natl Acad Sci USA 80:4045–4049
Lerman MI, Thayer RE, Singer MF (1983) Kpn I family of long interspersed repeated DNA sequences in primates: polymorphism of family members and evidence for transcription. Proc Natl Acad Sci USA 80:3966–3970
Maio JJ, Brown FL, McKenna WG, Musich PR (1981) Toward a molecular paleontology of primate genomes. II. The Kpn I families of alphoid DNAs. Chromosoma 83:127–144
Manuelidis L, (1980) Novel classes of mouse repeated DNAs. Nucleic Acids Res 8:3247–3258
Manuelidis L (1982a) Repeated DNA sequences and nuclear structure. In: Dover GA, Flavell RB (eds) Genome evolution. Academic Press, New York, pp 261–285
Manuelidis L (1982b) Nucleotide sequence definition of a major human repeated DNA, the HindIII 1.9-kb family. Nucleic Acids Res 10:3211–3219
Manuelidis L (1984a) Different CNS cell types display distinct and non-random arrangements of satellite DNA sequences. Proc Natl Acad Sci USA 81:3123–3127
Manuelidis L (1984b) Active nucleolus organizers are precisely positioned in adult central nervous system cells but not in neuroectodermal tumor cells. J Neuropathol Exp Neurol 43:225–241
Manuelidis L, Biro PA (1982) Genomic representation of the Hind-III 1.9-kb repeated DNA. Nucleic Acids Res 10:3221–3239
Manuelidis L, Manuelidis EE (1979) Surface growth characteristics of defined normal and neoplastic neuroectodermal cells in vitro. In: Zimmerman HM, (ed) Progress in neuropathology, vol. 4, Raven Press, New York, pp 235–285
Manuelidis L, Langer-Safer PR, Ward DC (1982) High resolution mapping of satellite DNA using biotin-labeled DNA probes. J Cell Biol 95:619–625
Marsden MPF, Laemmli UK (1979) Metaphase chromosome structure: evidence for a radial loop model. Cell 17:849–858
Martin SL, Voliva CF, Burton FH, Edgell MH, Hutchison CA III (1984) A large interspersed repeat found in mouse DNA contains a long open reading frame that evolves as if it encodes a protein. Proc Natl Acad Sci USA 81:2308–2312
Newman GR, Jasani B, Williams ED (1983a) The visualization of trace amounts of diaminobenzidine (DAB) polymer by a novel gold-sulphide-silver method. J Microsc 132:1–2
Newman GR, Jasani B, Williams ED (1983b) Metal compound intensification of the electron-density of diaminobenzidine. J Histochem Cytochem 31:1430–1434
Pan J, Elder JT, Duncan CH, Weissman SM (1981) Structural analysis of interspersed repetitive polymerase transcription units in human DNA. Nucleic Acids Res 9:1151–1170
Potter SS (1984) Rearranged sequences of a human Kpn I element. Proc Natl Acad Sci USA 81:1012–1016
Rubin CM, Houck CM, Deininger PL, Friedmann T, Schmid CW (1980) Partial nucleotide sequence of the 300 nucleotide interspersed repeated human DNA sequences. Nature 284:372–374
Sakonju S, Bogenhagen DF, Brown DD (1980) A control region in the center of the 5S RNA gene directs specific initiation of transcription. I. The 5′ border of the region. Cell 19:13–25
Schmeckpeper BJ, Willard HF, Smith KD (1981) Isolation and characterization of cloned human DNA fragments carrying reiterated sequences common to both autosomes and the X-chromosome. Nucleic Acids Res 9:1853–1872
Sedat J, Manuelidis L (1978) A direct approach to the structure of eukaryotic chromosomes. Cold Spring Harbor Symp Quant Biol 42:331–350
Shafit-Zagardo B, Maio JJ, Brown FL (1982) Kpn I families of long interspersed repetitive DNAs in human and other primate genomes. Nucleic Acids Res 10:3175–3179
Shafit-Zagardo B, Brown FL, Zavodny PJ, Maio JJ (1983) Transcription of the Kpn I families of long interspersed DNAs in human cells. Nature 304:277–280
Sharp PA (1983) Conversion of RNA to DNA in mammals: Alulike elements and pseudogenes. Nature 301:471–472
Singer MF (1982) Highly repeated sequences in mammalian genomes. Int Rev Cytol 76:67–112
Singer MF, Thayer RE, Grimaldi G, Fanning TG (1983) Homology between the Kpn I primate and Bam HI (MIF-1) rodent families of long interspersed repeated sequences. Nucleic Acids Res 11:5739–5745
Thayer RE, Singer ML (1983) Interruption of an α-satellite array by a short member of the Kpn I family of interspersed, highly repeated monkey DNA sequences. Mol Cell Biol 3:967–973
Varley KM, MacGregor HC, Euba HP (1980) Satellite DNA is transcribed on lampbrush chromosomes. Nature 283:686–688
Willard HF, Smith KD, Sutherland J (1983) Isolation and characterization of a major tandem repeat family from the human X chromosome. Nucleic Acids Res 11:2017–2033
Wu JC, Manuelidis L (1980) Sequence definition and organization of a human repeated DNA. J Mol Biol 142:363–386
Yang TP, Hansen SK, Oishi KK, Ryder DA, Hamkalo BA (1982) Characterization of a cloned repetitive DNA sequence concentrated on the human X chromosome. Proc Natl Acad Sci USA 79:6593–6597
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Manuelidis, L., Ward, D.C. Chromosomal and nuclear distribution of the HindIII 1.9-kb human DNA repeat segment. Chromosoma 91, 28–38 (1984). https://doi.org/10.1007/BF00286482
Received:
Revised:
Issue Date:
DOI: https://doi.org/10.1007/BF00286482