Abstract
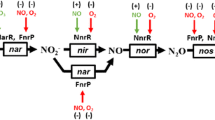
Operon fusion strains and mutants of Escherichia coli K-12 lacking the NADH-dependent nitrite reductase have been used to determine the regulation and physiological roles of two independent pathways for nitrite reduction to ammonia. Both the formate-and NADH-dependent pathways (Nrf and Nir, respectively) were totally repressed during aerobic growth, partially active during anaerobic growth in the absence of nitrite and further induced anaerobically by nitrite. Both were dependent upon a functional Fnr protein (a transcription activator of genes for anaerobic respiration). During anaerobic growth in the presence of nitrate, the Nir pathway was fully induced but Nrf was strongly repressed. Mutants defective in the NarL protein, which induces transcription of nitrate reductase genes but represses fumarate reductase genes in the presence of nitrate, were derepressed for Nrf activity during growth with nitrate, but the Nir enzyme was less active. The synthesis of Nrf components was also sensitive to glucose repression and weak activation by NarL during growth in the absence of nitrate. These data indicate that the Nir pathway provides a mechanism for detoxifying nitrite formed in the cytoplasm as a product of nitrate reduction. In contrast, the electrogenic reduction of nitrite by the Nrf pathway provides a secondary source of energy during anaerobic growth and is consequently repressed by the NarL protein when the thermodynamically more favourable electron acceptor, nitrate, is available. Two short DNA sequences, 5′-TACCAT-3′ and 5′-CTCCTT-3′, were found in the promoters of operons known to be activated or repressed by the NarL protein. It is proposed that NarL activates nir B transcription by binding to one or both of these sequences located 5′ to the RNA polymerase binding site, but represses other operons, including nrf, by binding close to the transcription start.
Similar content being viewed by others
Abbreviations
- Nir:
-
nitrite reduction by NADH
- Nrf:
-
nitrite reduction by formate
References
Abou-Jaoudé A, Chippaux M, Pascal M-C (1979a) Formate-nitrite reduction in Escherichia coli K 12 1. Physiological study of the system. Eur J Biochem 95: 309–314
Abou-Jaoudé A, Pascal M-C, Chippaux M (1979b) Formate-nitrite reduction in Escherichia coli K12 2. Identification of components involved in the electron transfer. Eur J Biochem 95: 315–321
Birkman A, Sawers RG, Bock A (1987) Involvement of the ntrA gene product in the anaerobic metabolism of Escherichia coli. Mol Gen Genet 210: 535–542
Chippaux M, Giudici D, Abou-Jaoudé A, Casse F, Pascal M-C (1978) Mutation leading to total lack of nitrite reductase activity in Escherichia coli. Mol Gen Genet 160: 225–229
Cole JA (1978) The rapid accumulation of large quantities of ammonia during nitrite reduction by Escherichia coli. FEMS Microbiol Lett 4: 327–329
Cole JA (1988) Assimilatory and dissimilatory reduction of nitrate to ammonia. In: Cole JA, Ferguson SJ (eds) The nitrogen and sulphur cycles. Symposium 42, Society for General Microbiology. Cambridge University Press, Cambridge, pp 281–329
Cole JA, Brown CM (1980) Nitrite reduction to ammonium by fermentative bacteria: a short circuit in the biological nitrogen cycle. FEMS Microbiol Lett 7: 65–72
Cole JA, Ward FB (1973) Nitrite reductase-deficient mutants of Escherichia coli K12. J Gen Microbiol 76: 21–29
Cotter PA, Gunsalus RP (1989) Oxygen, nitrate, and molybdenum regulation of dmsABC gene expression in Escherichia coli. J Bacteriol 171: 3817–3823
Drummond MH, Whitty PW, Wootton JC (1986) Sequence and domain relationships of ntrC and nifA from Klebsiella pneumoniae; homologies to other regulatory proteins. EMBO J 5: 441–447
Eiglmeier K, Honore N, Iuchi S, Lin ECC, Cole ST (1989) Molecular genetic analysis of FNR-dependent promoters. Mol Microbiol 3: 869–878
Fujita T, Sato R (1966a) Studies in soluble cytochromes in Enterobacteriaceae. III. Localization of cytochrome c 552 in the surface layer of cells. J Biochem 60: 568–577
Fujita T, Sato R (1966b) Studies on soluble cytochromes on Enterobacteriaceae. IV. Possible involvement of cytochrome c 552 in anaerobic nitrite metabolism. J Biochem 60: 691–700
Graham A, Boxer DH (1980) Arrangement of the respiratory nitrate reductase in the cytoplasmic membrane of Escherichia coli. FEBS Lett 113: 15–20
Graham A, Boxer DH (1981) The organisation of formate dehydrogenase in the cytoplasmic membrane of Escherichia coli. Biochem J 195: 629–637
Griffiths L, Cole JA (1987) Lack of redox control of the anaerobically-induced nirB + gene of Escherichia coli. Arch Microbiol 147: 364–369
Iuchi S, Lin ECC (1987) The narL gene product activates the nitrate reductase operon and represses the fumarate reductase and trimethylamine N-oxide reductase operons in Escherichia coli. Proc Natl Acad Sci USA 84: 3901–3905
Jackson RH, Cornish-Bowden A, Cole JA (1981) Prosthetic groups of the NADH-dependent nitrite reductase from Escherichia coli K12. Biochem J 193: 861–867
Jayaraman PS, Peakman TC, Busby SJW, Quincey RV, Cole JA (1987) Location and sequence of the promoter of the gene for the NADH-dependent nitrite reductase of Escherichia coli and its regulation by oxygen, the FNR protein and nitrite. J Mol Biol 196: 781–788
Kalman LV, Gunsalus RP (1989) Identification of a second gene involved in global regulation of fumarate reductase and other nitrate-controlled genes for anaerobic respiration in Escherichia coli. J Bacteriol 171: 3810–3816
Lambden PR, Guest JR (1976) Mutants of Escherichia coli K12 unable to use fumarate as an electron acceptor. J Gen Microbiol 97: 145–160
Li SF, DeMoss JA (1988) Location of sequences in the nar promoter of Escherichia coli necessary for regulation by Fnr and NarL. J Biol Chem 263: 13700–13705
MacGregor CH, Christopher AR (1978) Assymetric distribution of nitrate reductase sub-units in the cytoplasmic membrane of Escherichia coli. Arch Biochem Biophys 185: 204–213
Magasanik B (1988) Reversible phosphorylation of an enhancer binding protein regulates the transcription of bacterial nitrogen utilization genes. Trends Biochem Sci 13: 475–479
Motteram PAS, McCarthy JEG, Ferguson SJ, Jackson JB, Cole JA (1981) Energy conservation during the formate-dependent reduction of nitrite by Escherichia coli. FEMS Microbiol Lett 12: 317–320
Newman BM, Cole JA (1978) The chromosomal location and pleiotrophic effects of mutations in the nirA + gene of Escherichia coli K12: the essential role of nirA + in nitrite reduction and in other anaerobic redox reactions. J Gen Microbiol 106: 1–12
Nixon BT, Ronson CW, Ausubel FM (1986) Two-component regulatory systems responsive to environmental stimuli share strongly conserved domains with the nitrogen assimilation regulatory genes ntrB and ntrC. Proc Natl Acad Sci USA 83: 7850–7854
Nohno T, Noji S, Taniguchi S, Saito T (1989) The narL and narX genes encoding the nitrate-sensing regulators of Escherichia coli are homologous to a family of prokaryote two-component regulatory genes. Nucl Acid Res 17: 2947–2957
Pope NR, Cole JA (1982) Generation of a membrane potential by one of two independent pathways for nitrite reduction by Escherichia coli. J Gen Microbiol 128: 319–322
Pope NR, Cole JA (1984) Pyruvate and ethanol as electron donors for nitrite reduction Escherichia coli K12. J Gen Microbiol 130: 1279–1284
Stewart V (1982) Requirement of Fnr and NarL functions for expression of nitrate reductase expression in Escherichia coli K-12. J Bacteriol 151: 1320–1325
Stewart V, Berg BL (1988) Influence of nar (nitrate reductase) genes on nitrate inhibition of formate-hydrogen lyase and fumarate reductase gene expression in Escherichia coli K-12. J Bacteriol 170: 4437–4444
Stewart V, Parales J, Merkel SM (1989) Structure of genes narL and narX of the nar (nitrate reductase) locus in Escherichia coli K-12. J Bacteriol 171: 2229–2234
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Page, L., Griffiths, L. & Cole, J.A. Different physiological roles of two independent pathways for nitrite reduction to ammonia by enteric bacteria. Arch. Microbiol. 154, 349–354 (1990). https://doi.org/10.1007/BF00276530
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/BF00276530