Summary
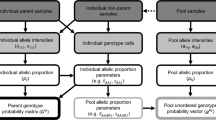
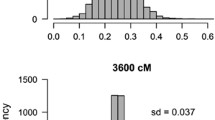
There has recently been a burgeoning interest in the analysis of paternity patterns for natural populations because of its relevance to population genetic phenomena such as the distance between successful mates, relative male reproductive success and gene flow. In this paper we develop a method of analyzing populational patterns of paternity, the fractional paternity method, and compare its performance to two other commonly used methods of paternity analysis (simple exclusion and the most-likely methods). We show that the fractional method is the most accurate method for determining populational patterns of paternity because it assigns paternity to all progeny examined, and because it avoids biases inherent in the other paternity analysis methods when model assumptions are met. In particular, it avoids a systematic bias of the most-likely paternity assignment method, which has a tendency to over-assign paternity of progeny to certain male parents with a greater than average number of homozygous marker loci. We also demonstrate the effect of linkage of some of the marker loci on paternity assignment, showing how the knowledge of the linkage phase of male and female parents in the population can significantly improve the accuracy of the estimates of populational patterns of paternity. Knowledge of the linkage phase of individuals in a population is usually unknown and difficult to assess without progeny testing, which involves considerable labor. However, we show how the linkage phase of hermaphroditic individuals in a population can be obtained in conjunction with the paternity analysis if progeny can be obtained from each hermaphroditic individual in the population, thereby avoiding the problem of traditional progeny testing. Applications of the fractional paternity approach developed herein should contribute significantly to our understanding of the mating patterns in, and hence the evolution of, natural populations.
Similar content being viewed by others
References
Brown AHD, Barrett SCH, Morgan GF (1985) Mating system estimation in forest trees: models, methods and meanings. In: Gregorius HR (ed) Population Genetics in Forestry. Springer, Berlin Heidelberg New York pp 32–49
Brown AHD, Grant JE, Pullen R (1986) Outcrossing and paternity in Glycine argyrea by pair fruit analysis. Biol J Linn Soc 29:283–294
Chakraborty R, Hedrick PW (1983) Paternity exclusion and the paternity index for two linked loci. Hum Hered 33:12–23
Chakraborty R, Shaw M, Schull WJ (1974) Exclusion of paternity: the current state of the art. Am J Hum Genet 26:477–488
Cheliak WM, Skroppa T, Pitel JA (1987) Genetics of the polycross. I. Experimental results from Norway spruce. Theor Appl Genet 73:321–329
Devlin B, Stephenson AG (1987) Sexual variations among plants of a perfect-flowered species. Am Nat 130:199–218
Elandt-Johnson RC (1971) Probability models and statistical methods in genetics. Wiley, New York
Ellstrand NC (1984) Multiple paternity within the fruits of the wild radish, Raphanus sativus. Am Nat 123:819–828
Ellstrand NC, Marshall DL (1985) Interpopulation gene flow by pollen in wild radish, Raphanus sativus. Am Nat 126:606–612
Ellstrand NC, Marshall DL (1986) Patterns of multiple paternity in populations of Raphanus sativus. Evolution 40:837–842
Ennos RA, Clegg MT (1982) Effect of population substructuring on the estimates of outcrossing rate in plant populations. Heredity 48:283–292
Ennos RA, Dodson RK (1987) Pollen success, functional gender and assortative mating in an experimental plant population. Heredity 58:119–126
Friedman ST, Adams WT (1985) Estimation of gene flow into two seed orchards of loblolly pines (Pinus taeda L.). Theor Appl Genet 69:609–615
Hamrick JL, Schnabel A (1985) Understanding the genetic structure of plant populations: some old problems and a new approach. In: Gregorius HR (ed) Population Genetics in Forestry. Springer, Berlin Heidelberg New York, pp 50–70
Handel SN (1983) Contrasting gene flow patterns and genetic subdivision in adjacent populations of Cucumis sativus (Cucurbitaceae). Evolution 37:760–771
Hanken J, Sherman PW (1981) Multiple paternity in Belding's ground squirrel litters. Science 212:351–353
Levin DA (1981) Dispersal versus gene flow in plants. Ann Mo Bot Gard 68:233–253
Levin DA (1983) An immigration-hybridization episode in Phlox. Evolution 37:575–582
Levin DA, Kerster HW (1974) Gene flow in seed plants. Evol Biol 7:139–220
Meagher TR (1986) Analysis of paternity within a natural population of Chamaelirium luteum. I. Identification of most-likely male parents. Am Nat 128:199–215
Meagher TR, Thompson EA (1986) The relationship between single parent and parent pair genetic likelihoods in genealogy reconstruction. Theor Popul Biol 29:87–106
Meagher TR, Thompson EA (1987) Analysis of parentage for naturally established seedlings within a population of Chamaelirium luteum (Liliaceae). Ecology 68:803–812
Muller-Starck G, Ziehe M (1984) Reproductive systems in conifer seed orchards. 3. Female and male fitnesses of individual clones realized in seeds of Pinus sylvestris L. Theor Appl Genet 69:173–177
Neale DB (1983) Population genetic structure of the shelterwood regeneration system in southwest Oregon. PhD thesis, Oregon State University, Corvallis
Neel JV, Schull WJ (1954) Human Heredity. University of Chicago Press, Chicago
Roeder KM, Devlin B, Lindsay BG (1988) Application of maximum likelihood methods to population genetic data for the estimation of individual fertilities. Biometrics (in press)
Ryman N, Chakraborty R (1982) Evaluation of paternity-testing data from the joint distribution of paternity index and the rate of exclusion. Hereditas 96:49–54
Schaal B (1980) Measurement of gene flow in Lupinus texensis. Nature 284:450–451
Schoen DJ, Stewart SC (1986) Variation in male reproductive investment and male reproductive success in White Spruce. Evolution 40:1109–1120
Selvin S (1980) Probability of nonpaternity determined by multiple allele codominant systems. Am J Hum Genet 32:276–278
Smith DB, Adams WT (1983) Measuring pollen contamination in clonal seed orchards with the aid of genetic markers. Proc 17th So Tree Improvement Conf, University of Georgia, Athens/GA, pp 69–77
Smouse PE, Chakraborty R (1986) The use of restriction fragment length polymorphism in paternity analysis. Am J Hum Genet 38:918–939
Thompson EA (1986) Likelihood inference of paternity. Am J Hum Genet 39:285–287
Trivers R (1985) Social Evolution. Benjamin/Cummings, Menlo Park
Valentin J (1980) Exclusion and attributions of paternity: practical experience of forensic genetics and statistics. Am J Hum Genet 32:420–431
Yampolsky E, Yampolsky H (1922) Distribution of sex forms in the phanerogamic flora. Bibliogr Genet 3:1–62
Author information
Authors and Affiliations
Additional information
Communicated by PMA Tigerstedt
Rights and permissions
About this article
Cite this article
Devlin, B., Roeder, K. & Ellstrand, N.C. Fractional paternity assignment: theoretical development and comparison to other methods. Theoret. Appl. Genetics 76, 369–380 (1988). https://doi.org/10.1007/BF00265336
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00265336