Summary
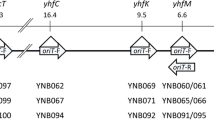
We cloned and sequenced a 402 by DNA segment containing the origin of conjugal transfer (oriT) of the IncW plasmid R388. Progressive deletions from each end of the sequence were assayed for oriT activity. Stepwise reductions in mobilization frequencies, representing the loss of functional elements, correlated with deletion of structural motifs in the sequence. A sequence of 330 by of oriT was sufficient for efficient mobilization. The first 86 by of the sequence contains five tandemly repeated DNA sequences of 11 bp, followed by a 10 by perfect inverted repeat. Deletion of the first 95 by reduced the frequency of transfer by a hundred-fold. The sequence between by 183 and 218 was necessary and sufficient for low frequency mobilization and, thus, it was assumed to contain the nick site. This basis core was cloned as a 60 by segment (from by 176–236) that could be mobilized at low frequency. It includes two inverted repeats and a perfect integration host factor (IHF) consensus binding site. A third functionally important segment in oriT was located between by 260 and 330. The DNA sequence of the oriT of R388 could be aligned with that of the broad-host-range IncN plasmid R46. Moreover, the relative positions of the three inverted repeats are also conserved. Overall sequence similarity was 52%, but was significantly higher in particular regions, whch coincided with the functionally important segments mapped by deletion analysis. Conservation of these segments provided independent support for their essential role in oriT function.
Similar content being viewed by others
References
Avila P, de la Cruz F (1988) Physical and genetic map of the IncW plasmid R388. Plasmid 20:155–157
Bartolomé B, Jubete Y, Martínez E, de la Cruz F (1991) Construction and properties of a family of pACYC184-derived cloning vectors, compatible with pBR322 and its derivatives. Gene, in press
Bastia D (1978) Determination of restriction sites and the nucleotide sequence surrounding the relaxation site of ColE1. J Mol Biol 124:601–639
Bernardi A, Bernardi F (1984) Complete sequence of pSC101. Nucleic Acids Res 12:9415–9426
Bolland S, Llosa M, Avila P, de la Cruz F (1990) General organization of the conjugal transfer genes of the IneW plasmid R388, and interactions between R388 and IncN and IncP plasmids. J Bacteriol 172:5795–5802
Bradley DE, Taylor DE, Cohen DR (1980) Specification of surface mating systems among conjugative drug resistance plasmids in Escherichia coli K-12. J Bacteriol 143:1466–1470
Brasch MA, Meyer RJ (1987) A 38 base-pair segment of DNA is required in cis for conjugative mobilization of broad hostrange plasmid R1162. J Mol Biol 198: 361–369
Broome-Smith J (1980) RecA independent, site-specific recombination between ColEl or ColK and a miniplasmid they complement for mobilization and relaxation: implications for the mechanism of DNA transfer during mobilization. Plasmid 4:51–63
Brown AMC, Willetts NS (1981) A physical and genetic map of the IncN plasmid R46. Plasmid 5:188–201
Buchanan-Wollaston V, Passiatore JE, Cannon F (1987) The mob and oriT mobilization functions of a bacterial plasmid promote its transfer to plants. Nature 328:172–175
de la Campa AG, del Solar GH, Espinosa M (1990) Initiation of replication of plasmid pLS1. The initiator protein RepB acts on two distant DNA regions. J Mol Biol 213:247–262
Campbell JL, Richards CC, Studier FW (1978) Genetic recombination and complementation between bacteriophage T7 and cloned fragments of T7-DNA. Proc Natl Acad Sci USA 75:2276–2280
Chung CT, Miller RH (1988) A rapid and convenient method for the preparation and storage of competent bacterial cells. Nucleic Acids Res 16:3580
Clark AJ, Warren GJ (1979) Conjugal transmission of plasmids. Annu Rev Genet 13:99–125
Coupland GM, Brown AMC, Willetts NS (1987) The origin of transfer (oriT) of the conjugative plasmid R46: Characterization by deletion analysis and DNA sequencing. Mol Gen Genet 208:219–225
de la Cruz F, Grinsted J (1982)Genetic and molecular characterization of Tn21, a multiple resistance transposon from R100.1. J Bacteriol 151:222–228
Datta N, Hedges RW (1972) Trimethoprim resistance conferred by W plasmid in Enterobacteriaceae. J Gen Microbiol 72:349–355
Derbyshire KM, Willetts NS (1987) Mobilization of the non-conjugative plasmid RSF1010: A genetic analysis of its origin of transfer. Mol Gen Genet 206:154–160
Drolet M, Zanga P, Lau PCK (1990) The mobilization and origin of transfer regions of a Thiobacillus ferrooxidans plasmid: relatedness to plasmids RSF1010 and pSC101. Mol Microbiol 4:1381–1391
Falkow S, Guerry P, Hedges RW, Datta N (1974) Polynucleotide sequence relationships among plasmids of the I compatibility complex. J Gen Microbiol 85:65–76
Finlay BB, Frost LS, Paranchych W (1986) Origin of transfer of IncF plasmids and nucleotide sequences of the Type II oriT, traM, and traY alleles from ColB4-K98 and the Type IV traY allele from R100-1. J Bacteriol 168:132–139
Finnegan J, Sherratt D (1982) Plasmid ColE1 conjugal mobility: the nature of bom, a region required in cis for transfer. Mol Gen Genet 185:344–351
Fishel RA, James AA, Kolonder R (1981) recA-independent general genetic recombination of plasmids. Nature 294:184–186
Friden P, Voelkel K, Sternglanz R, Freundlich M (1984) Reduced expression of the isoleucine and valine enzymes in integration host factor mutants of Escherichia coli. J Mol Biol 172:573–579
Fürste JP, Ziegelin G, Pansegrau W, Lanka E (1987) Conjugative transfer of promiscuous plasmid RP4: plasmid specified functions essential for formation of relaxosomes. In: McMacken R, Kelly TJ (eds) DNA Replication and Recombination. UCLA Symp Mol Cell Biol, New Series vol 47. Alan R. Liss, New York, pp 553–564
Fürste JP, Pansegrau W, Ziegelin G, Kröger M, Lanka E (1989) Conjugative transfer of promiscuous IncP plasmids: Interaction of plasmid-encoded products with the transfer origin. Proc Natl Acad Sci USA 86:1771–1775
Göldner A, Graus H, Högenauer G (1987) The origin of transfer of P307. Plasmid 18:76–83
Goto N, Shoji A, Horiuchi S, Nakaya R (1984) Conduction of nonconjugative plasmids by F′lac is not necessarily associated with transposition of the γδ sequence. J Bacteriol 159:590–596
Guiney DG, Lanka E (1989) Conjugative transfer of IncP plasmids. In: Thomas CM (ed) Promiscuous plasmids of Gram-negative bacteria. Academic Press, London, pp 27–56
Guiney DG, Yacobson E (1983) Location and nucleotide sequence of the transfer origin of the broad host range plasmid RK2. Proc Natl Acad Sci USA 80:3595–3598
Guiney DG, Deiss C, Simnad V (1988) Location of the Relaxation Complex Nick Site within the Minimal Origin of Transfer Region of RK2. Plasmid 20:259–265
Hanahan D (1985) Techniques for transformation of E. coli. In: Glover DM (ed) DNA cloninig: a Practical Approach, vol 1. IRL Press, Oxford, pp 109–135
Henikoff S (1984) Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene 28:351–359
Ippen-Ihler KA (1986) The conjugation system of F, the fertility factor of Escherichia coli. Annu Rev Genet 20:593–624
Jacob AE, Shapiro JA, Yamamoto L, Smith DI, Cohen SN, Berg D (1977) Plasmids studied in Escherichia coli and other enteric bacteria. In: Bukhari AI, Shapiro TA, Adhya SL (eds) DNA insertion elements, plasmids and episomes. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 635–636
Koepsel RR, Khan SA (1986) Static and initiator protein-enhanced bending of DNA at a replication origin. Science 233:1316–1318
Komano T, Toyoshima A, Morita K, Nisioka T (1988) Cloning and Nucleotide Sequence of the oriT Region of the IncIl Plasmid R64. J Bacteriol 170:4385–4387
Lahue EE, Matson SW (1990) Purificed Escherichia coli F-Factor TraY protein binds oriT. J Bacteriol 172:1385–1391
Leong JM, Nunes-Duby S, Lesser CF, Youderian P, Susskind MM, Landy A (1985) The psi80 and p22 attachment sites. Primary structure and interaction with Escherichia coli integration host factor. J Biol Chem 260:4468–4477
Makris JC, Nordmann PL, Reznikoff WS (1990) Integration Host Factor plays a role in IS50 and Tn5 transposition. J Bacteriol 172:1368–1373
Maniatis T, Fritsch EF, Sambrook J (1982) Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
Martinez E, de la Cruz F (1988) Transposon Tn21 encodes a recAindependent site-specific integration system. Mol Gen Genet 211:320–325
McIntire SA, Dempsey WB (1987) oriT sequence of the antibiotic resistance plasmid R100. J Bacteriol 169:3829–3832
Mukherjee S, Patel I, Bastia D (1985) Conformational changes in a replication origin induced by an initiator protein. Cell 43:189–197
Nordheim A (1979) Charakterisierung von bakteriellen Plasmid DNA-Protein Relaxationskomplexen. PhD thesis, Freie Universität, Berlin, FRG
Olsen RH, Siak JS, Gray RH (1974) Characteristics of PRD1, a plasmid-dependent broad host range DNA bacteriophage. J Virol 14:689–699
Ostermann E, Kricek F, Högenauer G (1984) Cloning the origin of transfer region of the resistance plasmid R1. EMBO J 3:1731–1735
Pansegrau W, Ziegelin G, Lanka E (1988) The origin of conjugative IncP plasmid transfer: interaction with plasmid-encoded products and the nucleotide sequence at the relaxation site. Biochim Biophys Acta 951:365–374
Pérez-Martin J, del Solar GH, de la Campa AG, Espinosa M (1988) Three regions in the DNA of plasmid pLS1 show sequencedirected static bending. Nucleic Acids Res 16:9113–9126
Roessler E, Fenwick RG Jr, Chinault AC (1985) Analysis of mobilization elements in plasmids from Shigella flexneri. J Bacteriol 161:1233–1235
Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-termination inhibitors. Proc Natl Acad Sci USA 74:5463–5467
Smith GR (1989) Homologous recombination in E. coli: multipe pathways for multiple reasons. Cell 58:807–809
Snijders A, van Patten AJ, Veltkamp E, Nijkamp HJJ (1983) Localization and nucleotide sequence of the bom region of Clo DF13. Mol Gen Genet 192:444–451
Stewart GSAB, Lubinsky-Mink S, Jackson CG, Cassel A, Kuhn J (1986) pHG165: A pBR322 Copy Number Derivative of pUC8 for Cloning and Expression. Plasmid 15:172–181
Stockes HW, Hall RM (1989) A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol Microbiol 3:1669–1683
Thompson R, Taylor L, Kelly K, Everett R, Willetts N (1984) The F plasmid origin of transfer: DNA sequence of wild-type and mutant origins and location of origin-specific nicks. EMBO 73:1175–1180
Thompson TL, Centola MB, Deonier RC (1989) Location of the nick at oriT of the F plasmid. J Mol Biol 207:505–512
Tsai MM, Fu YHF, Deonier RC (1990) Intrinsic bends and Integration Host Factor binding at F plasmid oriT. J Bacteriol 172:4603–4609
Valentine CRI, Kado CI (1989) Molecular genetics of IncW plasmids. In: Thomas CM (ed) Promiscuous plasmids of Gramnegative bacteria. Academic Press, London, pp 125–163
Vieira J, Messing J (1982) The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19:259–268
Wackernagel W (1973) Genetic transformation in E. coli: the inhibitory role of the recBC DNAse. Biochem Biophys Res Commun 51:306–311
Wang JC, Giaever GN (1988) Action at a distance along a DNA. Science 240:300–304
Warren GJ, Clark AJ (1980) Sequence-specific recombination of plasmid ColEl. Proc Natl Acad Sci USA 77:6724–6728
Willetts N, Wilkins B (1984) Processing of Plasmid DNA During Bacterial Conjugation. Microbiol Rev 48:24–41
Winans SC, Walker GC (1985) Conjugal transfer system of the IncN plasmid pKM101. J Bacteriol 161:402–410
Yamamoto T, Motegi A, Takei T, Okayama H, Sawai T (1984) Plasmid R46 provides a function that promotes recA-indepen-dent deletion, fusion and resolution of replication. Mot Gen Genet 193:255–262
Ziegelin G, Füirste JP, Lanka E (1989) TraJ protein of plasmid RP4 binds to a 19-base pair invert sequence repetition within the transfer origin. J Biol Chem 264:11989–11994
Author information
Authors and Affiliations
Additional information
Communicated by H. Hennecke
Rights and permissions
About this article
Cite this article
Llosa, M., Bolland, S. & de la Cruz, F. Structural and functional analysis of the origin of conjugal transfer of the broad-host-range IneW plasmid R388 and comparison with the related IncN plasmid R46. Molec. Gen. Genet. 226, 473–483 (1991). https://doi.org/10.1007/BF00260661
Issue Date:
DOI: https://doi.org/10.1007/BF00260661