Summary
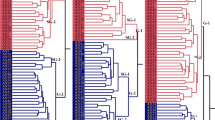
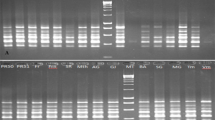
The objectives of this study were to assess the degree of restriction fragment length polymorphism (RFLP) in Cucumis melo and to determine interrelationships among cultivated varieties. Initial screening of a genomic PstI library revealed that approximately 40% of the clones were repetitive. A total of 162 unique and low-copy sequence clones were hybridized to seven diverse accesions of C. melo and a C. sativus cultivar ‘Pacer’ to evaluate RFLP variation. Of these, 130 probes (80%) detected a polymorphism between C. melo accessions and C. sativus, and the majority were polymorphic with more than one enzyme digest. In contrast, only 53 probes (33%) were useful in differentiating at least one of the seven accessions. Of those, only 9% were informative with more than one enzyme digest. This indicates that within C. melo, the differences among accessions are due to infrequent base substitutions, whereas between the two species, differences are mainly due to genome rearrangements such as insertions and deletions or numerous base substitutions. Of the informative probes, 34 were used in analyzing 44 C. melo lines to establish a data base of RFLP hybridization patterns. Percent similarity based on RFLP profiles was computed among lines and analyzed by principal component analysis, to visualize relationships among lines. There were clear demarcations among, but not within, muskmelon and honeydew groups.
Similar content being viewed by others
References
Aviv H, Leder P (1972) Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci USA 69:1408–1414
Dane F (1983) Cucurbits. In: Tanksley SD, Orton TJ (eds) Isozymes in plant genetics and breeding, Part b. Elsevier, Amsterdam, pp 369–390
Deakin JR, Bohn GW, Whitaker TW (1971) Interspecific hybridization in Cucumis. Econ Bot 25:195–211
Feinberg AP, Vogelstein B (1983) A technique for radiolabelling DNA restriction endonuclease fragments to high specific activity. Anal Biochem 132:6
Feinberg AP, Vogelstein B (1984) A technique for radiolabelling DNA restriction endonuclease fragments to high specific activity, addendum. Anal Biochem 137:26
Figdore SS, Kenard W, Song KM, Slocum MK, Osborn TC (1988) Assessment of the degree of restriction fragment length polymorphism in Brassica. Theor Appl Genet 75:833–840
Helentjaris T, Burr B (eds) (1989) Development and application of molecular markers to problems in plant genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor/NY
Helentjaris T, King G, Slocum M, Siedenstrang C, Wegman S (1985) Restriction fragment polymorphisms as probes for plant diversity and their development as tools for applied plant breeding. Plant Mol Biol 5:109–118
Helentjaris T, Slocum M, Wright S, Schaefer A, Nienhuis J (1986) Construction of genetic linkage maps in maize and tomato using restriction fragment length polymorphisms. Theor Appl Genet 72:761–769
Ingle J, Timmis JN, Sinclair J (1975) The relationship between satellite deoxyribonucleic acid, ribosomal ribonucleic acid, gene redundancy, and genome size in plants. Plant Physiol 55:496–501
King G, Turner V, Hussey C, Wurtele E, Lee M (1988) Isolation and characterization of a tomato cDNA clone which codes for a salt-induced protein. Plant Mol Biol 10:401–412
Lichtenstein C, Draper J (1985) Genetic engineering of plants. In: Glover DM (ed) DNA cloning, vol II. A practical approach. IRL Press, Oxford Washington DC, pp 67–119
Maniatis T, Fritsch EF, Sambrook J (1982) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor/NY
Pearson GG, Timmis JN, Ingle J (1974) The differential replication of DNA during plant development. Chromosoma 45:281–294
Perl-Treves R, Zamir D, Navot N, Galun E (1985) Phylogeny of Cucumis based on isozyme variability and its comparison with plastome phylogeny. Theor Appl Genet 71:430–436
SAS (1985) SAS user's guide: statistics, Version 5. SAS Institute Inc., Cary/NC, pp 956
Shattuck-Eidens DS, Bell RN, Neuhausen SL, Helentjaris T (1990) DNA sequence variation within maize and melon: observations from PCR amplification and direct sequencing. Genetics 126:207–217
Soller M, Beckmann JS (1983) Genetic polymorphism in varietal identification and genetic improvement. Theor Appl Genet 67:25–33
Song KM, Osborn TC, Williams PH (1988) Brassica taxonomy based on nuclear restriction fragment length polymorphisms (RFLPs). Theor Appl Genet 76:593–600
Staub JE, Frederick L, Marty TC (1987) Electrophoretic variation in cross-compatible wild diploid species of Cucumis. Can J Bot 65:792–798
Tanksley SD (1983) Molecular markers in plant breeding. Plant Mol Biol Rep 1:3–8
Williams JGK, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18:6531–6535
Author information
Authors and Affiliations
Additional information
Communicated by J. Beckmann
Rights and permissions
About this article
Cite this article
Neuhausen, S.L. Evaluation of restriction fragment length polymorphism in Cucumis melo . Theoret. Appl. Genetics 83, 379–384 (1992). https://doi.org/10.1007/BF00224286
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00224286