Summary
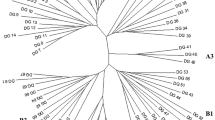
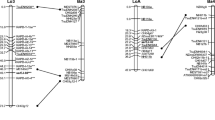
A detailed genetic linkage map of Brassica oleracea was constructed based on the segregation of 258 restriction fragment length polymorphism loci in a broccoli × cabbage F2 population. The genetic markers defined nine linkage groups, covering 820 recombination units. A majority of the informative genomic DNA probes hybridized to more than two restriction fragments in the F2 population. “Duplicate” sequences having restriction fragment length polymorphism were generally found to be unlinked for any given probe. Many of these duplicated loci were clustered non-randomly on certain pairs of linkage groups, and conservation of the relative linkage arrangement of the loci between linkage groups was observed. While these data support previous cytological evidence for the existence of duplicated regions and the evolution of B. oleracea from a lower chromosome number progenitor, no evidence was provided for the current existence of blocks of homoeology spanning entire pairs of linkage groups. The arrangement of the analyzed duplicated loci suggests that a fairly high degree of genetic rearrangement has occurred in the evolution of B. oleracea. Several probes used in this study were useful in detecting rearrangements between the B. oleracea accessions used as parents, indicating that genetic rearrangements have occurred in the relatively recent evolution of this species.
Similar content being viewed by others
References
Armstrong KC, Keller WK (1982) Chromosome pairing in haploids of Brassica oleracea. Can J Genet Cytol 24:735–739
Arus P, Orton TJ (1983) Inheritance and linkage relationships of isozyme loci in Brassica oleracea. J Hered 74:405–412
Bernatsky R, Tanksley S (1986) Toward a saturated linkage map in tomato based on isozymes and random cDNA sequences. Genetics 112:887–898
Bonierbale MW, Plaisted RL, Tanksley SD (1988) RFLP maps based on a common set of clones reveals modes of chromosomal evolution in potato and tomato. Genetics 120:1095–1103
Burr B, Evola SV, Burr FA, Beckmann JS (1983) The application of restriction fragment length polymorphism to plant breeding. In: Setlow J, Hollaender A (eds) Genetic engineering: principles and methods, Vol 5. Plenum Press, New York, pp 45–59
Chang C, Bowman J, DeJohn J, Lander E, Meyerowitz E (1988) Restriction fragment length polymorphism linkage map for Arabidopsis thaliana. Proc Natl Acad Sci USA 85:6856–6860
Figdore SS, Kennard WC, Song KM, Slocum MK, Osborn TC (1988) Assessment of the degree of restriction fragment length polymorphism in Brassica. Theor Appl Genet 75:833–940
Gottlieb LD (1983) Isozyme number and phylogeny. In: Jensen U, Fairbrothers DE (eds) Proteins and nucleic acids in plant systemics. Springer, Berlin Heidelberg New York pp 209–221 (Proceedings life sciences)
Gustafsson M, Bentzer B, Bothmer R von, Snogerup S (1976) Meiosis in Greek Brassica of the oleracea group. Bot Not 129:73–84
Haga T (1938) Relationships of genome to secondary pairing in Brassica. Jpn J Genet 13:277–284
Havey MJ, Muehlbauer FJ (1989) Linkages between restriction fragment length, isozyme, and morphological markers in lentil. Theor Appl Genet 77:395–401
Helentjaris T, Gesteland R (1983) Evaluation of random cDNA clones as probes for human restriction fragment polymorphisms. J Mol Appl Genet 2:237–247
Helentjaris T, King G, Slocum M, Siedenstrang C, Wegman S (1985) Restriction fragment polymorphisms as probes for plant diversity and their development as tools for applied plant breeding. Plant Mol Biol 5:109–118
Helentjaris T, Slocum M, Wright S, Schaefer A, Nienhuis J (1986) Construction of genetic linkage maps in maize and tomato using restriction fragment length polymorphisms. Theor Appl Genet 72:761–766
Helentjaris T, Weber D, Wright S (1988) Identification of the genomic locations of duplicated sequences in maize by analysis of restriction fragment length polymorphisms. Genetics 118:353–363
Landry BS, Kesseli RV, Farrara B, Michelmore R (1987) Agenetic map of lettuce (Lactuca sativa L.) with restriction fragment length polymorphism, isozyme, disease resistance and morphological markers. Genetics 116:331–337
McCouch SR, Kochert G, Yu ZH, Wang ZY, Khush GS, Coffman WR, Tanksley SD (1988) Molecular mapping of rice chromosomes. Theor Appl Genet 76:815–829
Ohno S (1970) Evolution by gene duplication. Springer, Berlin Heidelberg New York
Osborn TC, Alexander DC, Fobes JF (1987) Identification of restriction fragment length polymorphisms linked to genes controlling soluble solids content in tomato fruit. Theor Appl Genet 73:350–356
Prakash S, Hinata K (1980) Taxonomy, cytogenetics, and origin of crop Brassica, a review. Opera Bot 55:1–59
Quiros CF, Ochoa O, Kianian SF, Douches D (1987) Analysis of the Brassica oleracea genome by the generation of B. campestris-oleracea chromosome addition lines: characterization by isozyme and rDNA genes. Theor Appl Genet 74:758–766
Quiros CF, Ochoa O, Douches D (1988) Exploring the role of x=7 species in Brassica evolution: Hybridization with B. nigra and B. oleracea. J Hered 79:351–358
Robbelen G (1960) Beiträge zur Analyse des Brassica-Genomes. Chromosoma 2:205–228
Sampson DR (1978) A second gene for hairs in B. oleracea and its tentative location in linkage group four. Can J Genet Cytol 20:101–109
Song KM, Osborn TC, Williams PH (1988a) Brassica taxonomy based on nuclear restriction fragment length polymorphisms (RFLPs). 1. Genome evolution of the diploid and amphidiploid species. Theor Appl Genet 75:784–794
Song KM, Osborn TC, Williams PH (1988b) Brassica taxonomy based on nuclear restriction fragment length polymorphisms (RFLPs). 2. Preliminary analysis of subspecies within B. rapa (syn. campestris) and B. oleracea. Theor Appl Genet 76:593–600
Tanksley SD, Pichersky E (1988) Organization and evolution of sequence in the plant nuclear genome. In: Gottlieb LD, Jain SK (eds) Plant evolutionary biology. Chapman and Hall, New York, pp 55–83
Tanksley SD, Bernatsky R, Lapitan NL, Prince JP (1988) Conservation of gene repertoire but not gene order in pepper and tomato. Proc Natl Acad Sci USA 85:6419–6423
Wills AB (1977) A preliminary gene list in Brassica oleracea. Eucarpia Cruciferae News 2:22–24
Yarnell SH (1956) Cytogenetics of the vegetable crops. II. Crucifers. Bot Rev 22:81–166
Author information
Authors and Affiliations
Additional information
Communicated by J. Beckmann
Rights and permissions
About this article
Cite this article
Slocum, M.K., Figdore, S.S., Kennard, W.C. et al. Linkage arrangement of restriction fragment length polymorphism loci in Brassica oleracea . Theoret. Appl. Genetics 80, 57–64 (1990). https://doi.org/10.1007/BF00224016
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00224016