Abstract
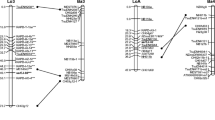
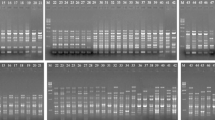
Two genomic maps were constructed for one individual tree of maritime pine, Pinus pinaster Ait., using a common set of 263 RAPD markers (random amplified polymorphic DNA). The RAPD markers were chosen from a larger number of polymorphic RAPD fragments on the basis of repeatability and inheritance in a three-generation pedigree. The maps were constructed from two independent mapping samples of 62 megagametophytes (In) from a self cross and from an open-pollinated cross. The markers were grouped (LOD≥4; θ≤0.25) and assigned to 13 major and 5 minor linkage groups. Two framework maps were constructed using the ordering criterion of interval support≥3. Comparison of the two framework maps suggested that the locus order was incorrect for 2% of the framework markers. A bootstrap analysis showed that this error rate was representative for our data set. The results showed that framework maps constructed using RAPD markers were repeatable and that differences in locus order for maps of different genotypes or species could result from chance. The total map distance was 1380 cM, and the map provided coverage of approximately 90% of the genome.
Similar content being viewed by others
References
Bahrman N, Damerval C (1989) Linkage relationships of loci controlling protein amounts in maritime pine (Pinus pinaster Ait.). Heredity 63:267–274
Beavis WD, Grant D (1991) A linkage map based on information from four F2 populations of maize, Zea mays L. Theor Appl Gen-et 82:636–644
Bonierbale MW, Plaisted RL, Tanksley SD (1988) RFLP maps based on a common set clones reveal modes of chromosomal evolution in potato and tomato. Genetics 120:1095–1103
Bradshaw HD, Stettler RF (1994) Molecular genetics of growth and development in Populus II: Segregation distortion due to genetic load. Theor Appl Genet 89:551–556
Chakravarti A, Lasher LA, Reefer JE (1991) A maximum likelihood method for estimating genome length using genetic linkage data. Genetics 128:175–182
Darvasi A, Weinreb A, Minke V, Weller JI, Soller M (1993) Detecting marker-QTL linkage and estimating QTL gene effect and map location using a saturated genetic map. Genetics 134:943–951
Devey ME, Fiddler TA, Liu BH, Knapp SJ, Neale BD (1994) An RFLP linkage map for loblolly pine based on a three-generation outbred pedigree. Theor Appl Genet 88:273–278
Doyle JJ, Doyle JL (1987) Isolation of DNA from fresh plant tissue. Focus 12:13–15
Efron B (1982) The jackknife, the bootstrap and other resampling plans. Society for Industrial and Applied Mathematics, Philadelphia, Pa.
Gebhardt C, Ritter E, Barone A, Debener T, Walkemeier B, Schachtschabel U, Kaufmann H, Thompson RD, Bonierbale MW, Ganal MW, Tanksley SD, Salamini F (1991) RFLP maps of potato and their alignment with the homeologous tomato genome. Theor Appl Genet 83:49–57
Gerber S, Rodolphe F, Bahrman N, Baradat P (1993) Seed-protein variation in maritime pine (Pinus pinaster Ait.) revealed by twodimensional electrophoresis: genetic determinism and construction of a linkage map. Theor Appl Genet 85:521–528
Grattapaglia D, Sederoff RR (1994) Genetic linkage maps of Eucalyptus grandis and E. urophylla using a pseudo-testcross mapping strategy and RAPD markers. Genetics 137:1121–1137
Groover A, Devey M, Fiddler T, Lee J, Megraw R, Mitchell-Olds T, Sherman B, Vujcic S, Williams C, Neale D (1994) Identification of quantitative trait loci influencing wood specific gravity in an outbred pedigree of loblolly pine. Genetics 138:1293–1300
Higgins MJ, Turnel C, Noolandi J, Neumann PE, Lalande M (1990) Construction of the physical map for three loci in chromosome band 13ql4: Comparison to the genetic map. Proc Natl Acad Sci USA 87:3415–3419
Hulbert SH, Ilott TW, Legg EJ, Lincoln SE, Lander ES, Michelmore RW (1987) Genetic analysis of the fungus, Bremia lactucae, using restriction length polymorphism. Genetics 120:947–958
Keats BJS, Sherman SL, Morton NE, Robson EB, Buetow KH, Cann HM, Cartwright PE, Chakravarti A, Francke U, Green PP, Ott J (1991) Guidelines for human linkage maps: an international system for human linkage maps (ISLM, 1990). Genomics 9:557–560
Kesseli RV, Paran I, Michelmore RW (1994) Analysis of a detailed genetic linkage map of Lactuca sativa (Lettuce) constructed from RFLP and RAPD markers. Genetics 136:1435–1446
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Lander ES, Botstein D (1989) Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121:185–199
Lange K, Boehnke M (1982) How many polymorphic genes will it take to span the human genome? Am J Hum Genet 34:842–845
Liu BH, Knapp SJ (1992) GMENDEL 2.0, a software for gene mapping. Oregon State University, Corvallis, Ore.
Moran GF, Bell JC, Hilliker AJ (1983) Greater meiotic recombination in male vs. female gametes in Pinus radiata. J Hered 74:62
Neale DB, Williams CG (1991) Restriction fragment length polymorphism mapping in conifers and applications to forest genetics and tree improvement. Can J For Res 21:545–554
Nelson CD, Nance WL, Doudrick RL (1993) A partial genetic linkage map of slash pine, Pinus elliottii Englem var ‘elliottii’ based on random amplified polymorphic DNA's. Theor Appl Genet 87:145–151
O'Malley DM, Guries RP, Nordheim EV (1986) Linkage analysis for 18 enzyme loci in Pinus rigida Mill. Theor Appl Genet 72:530–535
Ott J (1991) Analysis of human genetic linkage. Johns Hopkins University Press, Baltimore
Penner GA, Bush A, Wise R, Kim W, Domier L, Kasha K, Laroche A, Scoles G, Molnar SL, Fedak G (1993) Reproductibility of ranom amplified DNA, RAPD analysis among laboratories. PCR Methods Applic 2:341–345
Plomion C, Bahrman N, Durel CE, O'Malley DM (1995) Genomic mapping in maritime pine (Pinus pinaster) using RAPD and protein markers. Heredity 74 (in press)
Reiter RS, Williams J, Feldman K, Rafalski JA, Tingey SV, Scolnik PA (1992) Global and local genome mapping in Arabidopsis thalIana recombinant inbred lines and random amplified polymorphic DNAs. Proc Natl Acad Sci USA 89:1477–1481
Sax K, Sax HJ (1933) Chromosome number and morphology in the conifers. J Arnold Arbor Har Univ 14:356–375
Saylor LC, Smith BW (1966) Meiotic irregularity in species and intraspecific hybrids of Pinus. Am J Bot 53:453–468
Sorensen F (1967) Linkage between marker genes and embryonic lethal factors may cause disturbed segregation ratios. Silvae Genet 16:132–134
Sorensen TA (1948) A method of establishing groups of equal amplitude in plant sociology based on similarity of species content. Biol Skr K Danske Vidensk Selsk 5:1–34
Strauss SH, Conkle MT (1986) Segregation, linkage, and diversity of allozymes in knobcone pine. Theor Appl Genet 72:483–493
Tanksley SD, Bernatzky R, Lapitan NL, Prince JP (1988) Conservation of gene repertoire but not gene order in pepper and tomato. Proc Natl Acad Sci USA 85:6419–6423
Tanksley SD, Ganal MW, Prince JP, de Vicente MC, Bonierbale MW, Broun P, Fulton TM, Giovannoni JJ, Grandillo S, Martin GB, Messeguer R, Miller JC, Miller L, Paterson AH, Pineda O, Roder MS, Wing RA, Wu W, Young ND (1992) High-density molecular linkage maps of the tomato and potato genomes. Genetics 132:1141–1160
Tulsieram LK, Glaubitz JC, Kiss G, Carlson JE (1992) Single tree genetic linkage analysis in conifers using haploid DNA from megagametophytes. Bio/Technology 10:686–690
Whitkus R, Doebley J, Michael L (1992) Comparative genome mapping of sorghum and maize. Genetics 132:1119–1130
Williams JGK, Kubelik AR, Livak KJ, Rafalski JA (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18:6531–6535
Welsh J, McClelland M (1990) Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res 18:794–801
Author information
Authors and Affiliations
Additional information
Communicated by G. E. Hart
Rights and permissions
About this article
Cite this article
Plomion, C., O'Malley, D.M. & Durel, C.E. Genomic analysis in maritime pine (Pinus pinaster). Comparison of two RAPD maps using selfed and open-pollinated seeds of the same individual. Theoret. Appl. Genetics 90, 1028–1034 (1995). https://doi.org/10.1007/BF00222917
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00222917