Abstract
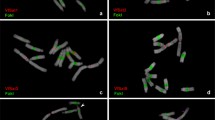
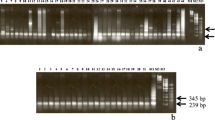
We report the cloning, sequencing and analysis of the major repetitive DNA of soybean (Glycine max). The repeat, SB92, was cloned as several monomers and trimers produced by digestion with XhoI. The deduced consensus sequence of the repeat is 92 base pairs long. Genomic sequences do not fluctuate in length. Their average homology to the consensus sequence is 92%. The consensus of SB92 contains slightly degenerated homologies for several 6-cutters. Therefore, many of them generate a ladder of 92-bp oligomers. The distribution of bands seems to be random, but the occurrence of sites for different 6-cutters varies widely. There is no obvious correlation between the sequences of the neighboring units of SB92 in cloned trimers. Also, there are none of the internal repetitive blocks reported for many satellite DNAs from other species. The SB92 repeat makes up 0.7% of total soybean DNA. This is equivalent to 8×104 copies, or 7 megabases. The repeat is organized in giant tandem blocks over 1 Mb in length, and there are fewer blocks than chromosomes. The polymorphism of these blocks is extremely high. The SB92 repeat is present in identical arrangement and number of copies in the ancestral subspecies Glycine soja. There are 10 times fewer copies of the repeat in a related species Vigna unguiculata (cowpea), and no homologies in several other more distant leguminous plants studied.
Similar content being viewed by others
References
Bassam BJ, Caetano-Anollés G, Gresshoff PM (1991) Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem 196:80–83
Bedbrook IR, Jones I, O'Dell M, Thompson RD, Flavell RB (1980) A molecular description of telomeric heterochromatin in Secale species. Cell 19:545–560
Benslimane AA, Dron M, Hartman C, Rode A (1986) Small tandemly repeated DNA sequences of higher plants likely originate from a tRNA ancestor. Nucleic Acids Res 14:8111–8119
Broun P, Ganal MW, Tanksley SD (1992) Telomeric repeats display high levels of heritable polymorphism among closely related plant varieties. Proc Natl Acad Sci USA 89:1354–1357
Doyle JJ (1988) 5S ribosomal gene variation in the soybean and its progenitor. Theor Appl Genet 75:621–624
Doyle JJ, Beachy RN (1985) Ribosomal gene variation in soybean and its relatives. Theor Appl Genet 70:369–376
Funke R, Kolchinsky A, Gresshoff PM (1993) Physical mapping of a region in the soybean genome containing duplicated sequences. Plant Mol Biol 22:437–446
Ganal M, Riede I, Hemleben V (1986) Organization and sequence analysis of two related satellite DNAs in cucumber (Cucumis sativus L.). J Mol Evol 23:23–30
Ganal MW, Broun P, Tanksley SD (1992) Genetic mapping of tandemly repeated telomeric DNA sequences in tomato. Genomics 14:444–448
Gill KS, Gill BS, Endo TR (1993) A chromosome region-specific mapping strategy reveals gene-rich telomeric ends in wheat. Chromosoma 102:374–381
Gurley WB, Hepburn AG, Key JL (1979) Sequence organization of the soybean genome. Biochim Biophys Acta 561:167–183
Ingham LD, Hanna WW, Baier JW, Hannah LC (1993) Origin of the main class of repetitive DNA within selected Pennisetum species. Mol Gen Genet 238:350–356
Kato I, Yakura K, Tanifugi S (1984) Sequence analysis of Vicia faba repeated DNA, the FokI element. Nucleic Acids Res 12:6415–6426
Keim P, Diers BW, Olson TC, Shoemaker RC (1990) RFLP mapping in soybean: association between marker loci and variation in quantitative traits. Genetics 126:735–742
Kolchinsky A, Gresshoff PM (1992) Nucleotide sequence of the 5S rRNA gene from Glycine soja. Plant Mol Biol 19:1045–1047
Kolchinsky A, Gresshoff PM (1993) Direct end-labeling of telomeres. Genome 36:224–229
Kolchinsky A, Gresshoff PM (1994) Plant telomeres as molecular markers. In: Gresshoff PM (ed) Plant genome analysis. CRC Press, Roca Baton, Fla., pp 117–124
Lark KG, Weisemann JM, Matthews BF (1993) A genetic map of soybean (Glycine max L.) using an intraspecific cross ‘Minsoy’ and ‘Noir 1’. Theor Appl Genet 86:901–906
Moore G, Gale MD, Kurata N, Flavell RB (1993) Molecular analysis of small grain cereal genomes: current status and prospects. Bio/Technology 11:584–589
Röder MS, Lapitan NLV, Sorrels ME, Tanksley SD (1993) Genetic and physical mapping of barley telomeres. Mol Gen Genet 238:294–303
Sambrook J, Fritsch EF, Maniatis T (1989). Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
Shoemaker RC, Guffy RD, Lorenzen LL, Specht JE (1992) Molecular genetic map in soybean: map utilization. Crop Sci 32:1091–1098
Singh RJ, Hymowitz T (1988) The genomic relationship between Glycine max (L.) Merr. and Glycine soja (Zieb. and Zucc.) as revealed by pachytene chromosome analysis. Theor Appl Genet 76:705–711
Vogt P (1992) Code domains in tandem repetitive DNA sequence structure. Chromosoma 101:585–589
Xia X, Erickson L (1993) An AT-rich satellite DNA sequence, E180, in alfalfa (Medicago sativa). Genome 36:427–432
Young ND, Fatokun CA, Danesh D, Menancio-Hautea D (1992) RFLP mapping in cowpea. In: Thottappilly G, Montin LM, Mohan Raj DR, Moore AW (eds) Biotechnology: enhancing research on tropical crops in Africa. CTA/IITA co-publication. IITA, Ibadan, Nigeria, pp 237–246
Author information
Authors and Affiliations
Additional information
Communicated by I. Potrkus
Rights and permissions
About this article
Cite this article
Kolchinsky, A., Gresshoff, P.M. A major satellite DNA of soybean is a 92-base pairs tandem repeat. Theoret. Appl. Genetics 90, 621–626 (1995). https://doi.org/10.1007/BF00222125
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00222125