Abstract
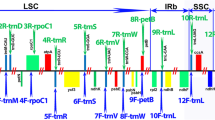
Chloroplast (cp) DNA from 32 genotypes representing eight genera and 19 species from the Andropogoneae tribe was analyzed using 15 restriction enzymes and Southern hybridization with 12 cpDNA probes that span the complete rice chloroplast genome. Six of the genera, Saccharum, Miscanthus, Erianthus, Narenga, Eccoilopus, and Sclerostachya, are part of the Saccharinae subtribe, whereas the other two, Zea and Sorghum, were used as outgroups. Narenga, Miscanthus, Erianthus, and Sclerostachya are presumed to have been involved in the evolution of Saccharum officinarum (“noble” or high sucrose sugarcane) via S. spontaneum and S. robustum. Southern hybridization with the rice cpDNA probes surveyed approximately 3% of the S. officinarum ‘Black Cheribon’ genome and yielded 62 restriction site mutations (18 informative) that were analyzed using cladistic parsimony and maximum likelihood. These site mutations placed the 32 genotypes into nine different chloroplast groups; seven from within the Saccharinae subtribe and the two outgroups (maize and Sorghum). Phylogenetic inferrence under various assumptions showed that the maternal lineages of Narenga, Miscanthus, Sclerostachya, and Saccharum formed a monophyletic group. This group displayed little variation. On the other hand, 5 of 6 Erianthus species and Eccoilopus longisetosus formed a separate group. The ‘Old World’ Erianthus/Eccoilopus chloroplast was very different from that of the rest of the ‘Saccharum complex’ members and was slightly more related to that of Sorghum bicolor. Placement of these Erianthus/Eccoilopus genotypes was, therefore, in conflict with analyses based on morphology. Surprisingly, Erianthus trinii, a New World species, had the same restriction sites as did one Miscanthus sinensis. One Miscanthus sp. from New Guinea that has a very high chromosome number (2n=192) had the same restriction sites as the majority of the Saccharum genus, suggesting that introgression between these genera occurs in the wild. The Saccharum genus was separated into two clades by single site mutation: one containing S. spontaneum, and the other containing all of the remaining Saccharum species and all 8 commerical hybrids (from various regions of the world). A physical map of the chloroplast of Saccharum officinarum ‘Black Cheribon’ was constructed using 5 restriction enzymes.
Similar content being viewed by others
References
Alberts VA, Mischler BD, Chase MW (1992) Charactertate weighing for restriction site data in phylogenetic reconstruction, with an example from chloroplast DNA. In: Soltis PS, Soltis DE, Doyle JJ (eds) Molecular systematics of plants. Chapman and Hall, New York, pp369–403
Al-Janabi SM, Honeycutt RJ, McClelland M, Sobral BWS (1993) A genetic linkage map of Saccharum spontaneum (L.) ‘SES 208’. Genetics 134:1249–1260
Artschwager E (1954) A taxonomic study of Saccharum sinense Roxb. and S. barberi Jeswiet. USDA Tech Bull No. 1089, Washington D.C.
Barbier P, Morishima H, Ishihama A (1991) Phylogenetic relationships of annual and perennial rice: probing by direct DNA sequencing. Theor Appl Genet 81:693–702
Bor NL (1960) The grasses of Burma, Ceylon, India and Pakistan. Pergamon Press, London
Bowman CM, Bonnard G, Dyer TA (1983) Chloroplast DNA variation between species of Triticum and Aegilops: location of the variation on the chloroplast genome and its relevance to the inheritance and classification of the cytoplasm. Theor Appl Genet 65:247–262
Brandes EW, Sartoris GB, Grassl CO (1939) Assembling and evaluating wild forms of sugarcane and closely related plants. Proc 6th Int Soc Sugarcane Technol Congr: 128–154
Bremer B (1991) Restriction site data from chloroplast DNA for phylogenetic reconstruction: is there only one accurate way of scoring? Plant Syst. Evol 175:39–54
Burner DM (1991) Cytogenetic analyses of sugarcane relatives (Andropogoneae: Saccharinae). Euphytica 54:125–133
Burnquist WB (1991) Development and application of restriction fragment length polymorphism technology in sugarcane (Saccharum spp.) breeding. PhD thesis, Cornell University, Ithaca N.Y.
Celarier RP (1956) Cytotaxonomy of the Andropogoneae. I. Subtribes Dimeriinae and Saccharinae. Cytologia 21:272–291
Clayton WD, Renvoize SA (1986) Genera Graminum: grasses of the world. Kew Bulletin Additional Series XIII, Her Majesty's Stationary Office, London
daSilva J, Sorrells ME, Burnquist WL, Tanksley SD (1993) Sugarcane (Saccharum spontaneum) genome analysis by means of restriction fragment length polymorphisms. Genome 36:782–791
Daniels J, Smith P, Paton N, Williams CA (1975) The origin of the genus Saccharum. Sugarcane Breed Newsl 36:24–39
DeBry RW, Slade NA (1985) Cladistic analysis of restriction endonuclease restriction maps within a maximum-likelihood framework. Syst Zool 34:21–34
Donoghue MJ, Sanderson MJ (1992) The suitability of molecular and morphological evidence in reconstructing plant phylogeny. In: Soltis PM, Soltis DE, Doyle JJ (eds) Molecular systematics of plants. Chapman and Hall, New York, pp 340–368
Duval MR, Doebley J (1990) Restriction site variation in the chloroplast genome of Sorghum (Poaceae). Syst Bot 15:472–480
Edwards A, Cavali-Sforza L (1963) The reconstruction of evolution. Ann Hum Genet 27:105
Farris JS (1970) Methods for computing Wagner trees. Syst Zool 34:21–34
Farris JS (1977) Phylogenetic analysis under Dollo's law. Syst. Zool 26:77–88
Feinberg AP, Vogelstein B (1983) A technique for radiolabelling DNA restriction fragments to a high specific activity. Anal Biochem 132:6–13
Felsenstein J (1973) Maximum likelihood and minimum-step methods for estimating evolutionary trees from data on discrete characters. Syst Zool 22:240–249
Felsenstein J (1989) PHYLIP-Phylogeny Inference Package. Cladistics 5:164–166
Glaszmann JC, Noyer JL, Fautret A, Feldmann P, Lanaud C (1989) Biochemical genetic markers in sugarcane. Theor Appl Genet 78:537–543
Glaszmann JC, Lu YH, Lanaud C (1990) Variation of nuclear ribosomal DNA in sugarcane. J Genet Breed 44:191–198
Gottlieb LD (1988) Towards molecular genetics in Clarkia: gene duplications and molecular characterization of PGI genes. Ann Mo Bot Gard 75:1169–1179
Grassl CO (1974) The origin of sugarcane. Sugarcane Breed Newsl 34:10–18
Grassl CO (1977) The origin of sugar-producing cultivars of Saccharum. Sugarcane Breed Newsl 39:8–33
Hartley W (1958) Studies on the origin, evolution, and distribution of the Gramineae. I. The tribe Andropogoneae. Aust J Bot 6:116–128
Hennig W (1965) Phylogenetic systematics. Annu Rev Entomol 10:97–116
Honeycutt RJ, BWS Sobral, P Keim, Irvine JE (1992) A rapid DNA extraction method for sugarcane and its relatives. Plant Mol Biol Rep 10:66–72
Keim P, Beavis W, Schupp J, Freestone R (1992) Evaluation of soybean RFLP marker diveristy in adapted germplasm. Theor Appl Genet 85:205–212
Keng YL (1957) Claves Generum et Specierum Graminearum Primarium Sinicarum. Peking
Kluge AG, Farris JS (1969) Quantitative phylogenetics and the evolution of anurans. Syst Zool 18:1–32
Manglesdorf AJ (1983) Cytoplasmic diveristy in relation to pests and pathogens. Sugarcane Breed Newsl 45:45–49
Maniatis T, Fritsch E, Sambrook J (1982) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
Mohan N, Sreenivasan TV (1983) Chromosome number in the genus Erianthus, Michx. (Poaceae) of Indonesian Arquipelago. Cell Chromosome Res 6:14–16
Moriya A (1990) List of chromosome numbers in the genus Saccharum and related genera. Jpn J Genet 16:126–136
Mukherjee SK (1957) Origin and distribution of Saccharum. Bot Gaz 119:55–61
Nair MK, Ratnambal MJ (1965) Pachytene analysis in Narenga x Sclerostachya hybrid. Proc 12th Int Soc Sugarcane Technol Congr:875–877
Palmer JD, Shields CR, Cohen DB, Orton TJ (1983) Chloroplast DNA evolution and the origin of amphidiploid Brassica species. Theor Appl Genet 65:181–189
Panje RR, Babu CN (1960) Studies in Saccharum spontaneum distribution and geographical association of chromosome numbers. Cytologia 25:152–172
Parthasarathy N (1953) Chromosome elimination in Saccharum. Nature 168:383–384
Price S (1957) Cytological studies in Saccharum and allied genera. III. Chromosome numbers in interspecific hybrids. Bot Gaz 144–159
Price S (1968) Chromosome transmission by Saccharum robustum in interspecific crosses. J Hered 59:245–247
Roach BT, Daniels J (1987) A review of the origin and improvement of sugarcane. In: Copersucar Int Sugarcane Breed Workshop. Copersucar, SP, Brazil, pp 1–32
Sankoff D (1975) Minimal mutation trees of sequences. Siam J Appl Math 28:35–42
Sankoff D, Morel C, Cederegen RJ (1983) Simultaneous comparison of three or more sequences related by a tree. In: Sankoff D, Krustal B (eds) Time warps, string edits, and macromolecules: the theory and practice of sequence comparisons. Addison-Wesley, Reading, Mass., pp 253–263
Shimada H, Suiguira M (1991) Fine structural features of the chloroplast genome: comparison of the sequenced chloroplast genomes. Nucleic Acids Res 19:983–995
Shimada H, Whittier RF, Hiratsuka J, Maeda Y, Hirai A, Sugiura M (1989) A physical map and clone bank of rice (Oryza sativa) chloroplast genome. Plant Mol Biol Rep 7:284–291
Soltis DE, Soltis PS, Milligan BG (1992) Intraspecific chloroplast variation: systematic and phylogenetic implications. In: Soltis PS, Soltis DE, Doyle JJ (eds) Molecular systematics of plants. Chapman and Hall, New York, pp 117–150
Stebbins GL (1956) Cytogenetics and evolution of the grass family. Am J Bot 43:890–905
Swofford DL (1991) PAUP: Phylogenetic analysis using parsimony, version 3.0s Computer program distributed by the Illinois Natural History Survey, Champaign, Ill.
Systma KJ, Smith JF, Berry PE (1991) Biogeography and evolution of morphology, breeding systems, flavonoids, and chloroplast DNA in the four Old World species of Fuchsia (Onagraceae). Syst Bot 16:257–269
Wilson MA, Gaut B, Clegg MT (1990) Chloroplast DNA evolves slowly in the palm family (Arecaceae). Mol Biol Evol 7:303–314
Wolfe KH, Sharp PM, Li WH (1989) Rates of synonomous substitution in plant nuclear genes. J Mol Evol 29:208–211
Author information
Authors and Affiliations
Additional information
Communicated by A. R. Hallauer
Rights and permissions
About this article
Cite this article
Sobral, B.W.S., Braga, D.P.V., LaHood, E.S. et al. Phylogenetic analysis of chloroplast restriction enzyme site mutations in the Saccharinae Griseb. subtribe of the Andropogoneae Dumort. tribe. Theoret. Appl. Genetics 87, 843–853 (1994). https://doi.org/10.1007/BF00221137
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00221137