Summary
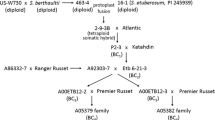
The location of sbm-1 on the Pisum sativum genetic map was determined by linkage analysis with eight syntenic molecular markers. Analysis of the progeny of two crosses confirmed that sbm-1 is on chromosome 6 and permitted a more detailed map of this chromosome to be constructed. The inclusion of Fed-1 and Prx-3 among the markers facilitated the comparison of our map with the classical genetic map of pea. The sbm-1 gene is most closely linked to RFLP marker GS185, being separated by a distance of about 8 cM. To determine the practical value of GS185 as a marker for sbm-1 in plant breeding programs, the GS185 hybridization pattern and virus-resistance phenotype were compared in of a collection of breeding lines and cultivars. Three GS185 hybridization patterns were discerned among the lines. A strong association was found between one of these patterns and resistance to PSbMV.
Similar content being viewed by others
References
Alconero R, Prowidenti R, Gonsalves D (1986) Three pea seedborne mosaic virus pathotypes from pea and lentil germ plasm. Plant Dis 70:783–786
Baggett JR, Hampton RO (1977) Oregon B422–15 and B445–66 pea seedborne mosaic virus-resistant breeding lines. Hort Science 12:506
Bentolila S, Guitton C, Bouvet N, Nykaza S, Freyssinet G (1991) Identification of an RFLP marker tightly linked to the Ht1 gene in maize. Theor Appl Genet 82:393–398
Diers BW, Mansur L, Imsande J, Shoemaker RC (1992) Mapping Phytophthora resistance loci in soybean with restriction fragment length polymorphism markers. Crop Sci 32:377–383
Feinberg A, Vogelstein B (1983) A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem 132:6–13
Gantt JS, Baldauf SL, Calie PJ, Weeden NF, Palmer JD (1991) Transfer of rpl22 to the nucleus greatly preceded its loss from the chloroplast and involved the gain of an intron. EMBO J 10:3073–3078
Gritton ET, Hagedorn DJ (1975) Linkage of the genes sbm and wlo in peas. Crop Sci 15:447–448
Hagedorn DJ, Gritton ET (1971) Registration of Wisconsin 7105 and 7106 pea germplasm. Crop Sci 11:945–946
Kraft JM, Giles RA (1978) Registration of VR74–410–2 and VR1492–1 pea germplasm. Crop Sci 18:1099
Kraft JM, Hampton RO (1980) Crop losses from pea seedborne mosaic virus in six processing pea cultivars. Plant Dis 64:922–924
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Lassner MW, Peterson P, Yoder JI (1989) Simultaneous amplification of multiple DNA fragments by polymerase chain reaction in the analysis of transgenic plants and their progeny. Plant Mol Biol Rep 7:116–128
Martin GB, Williams JGK, Tanksley SD (1992) Rapid identification of markers linked to a Pseudomonas resistance gene in tomato by using random primers and near-isogenic lines. Proc Natl Acad Sci USA 88:2336–2340
Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88:9828–9832
Muehlbauer FJ (1983) Eight germplasm lines of pea resistant to pea seedborne mosaic virus. Crop Sci 23:1019
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4326
Oberfelder R (1989) Immunoblotting: comparison of detection methods. BRL Focus 11:1–5
Paran I, Kesseli R, Michelmore R (1992) Identification of restriction fragment length polymorphism and random amplified polymorphic DNA markers linked to downy mildew resistance genes in lettuce, using near-isogenic lines. Genome 34:1021–1027
Polans NO, Folta KM, Elliot RC, Thompson WF (1991) Inheritance and linkage mapping of ferredoxin-1 in pea. J Hered 82:259–261
Provvidenti R (1990) Inheritance of resistance to pea mosaic virus in Pisum sativum. J Hered 81:143–145
Provvidenti R, Alconero R (1988a) Sources of resistance to pathotypes of pea seed-borne mosaic virus in the US plant introductions of Pisum sativum. Pisum Newslett 20:30–31
Provvidenti R, Alconero R (1988b) Inheritance of resistance to a lentil strain of pea seed-borne mosaic virus in Pisum sativum. J Hered 79:45–47
Provvidenti R, Alconero R (1988c) Inheritance of resistance to a third pathotype of pea seed-borne mosaic virus in Pisum sativum. J Hered 79:76–77
Provvidenti R, Muehlbauer FJ (1990) Evidence of a cluster of linked genes for resistance to pea seedborne mosaic virus and clover yellow vein virus on chromosome 6. Pisum Newslett 22:43–45
Reed KC, Mann DA (1985) Rapid transfer of DNA from agarose gels to nylon membranes. Nucleic Acids Res 13:7207–7221
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Cold Spring Harbor, New York
Skarzynska A (1988) Supplemental mapping data for chromosome 6. Pisum Newslett 20:34–36
Suiter KA, Wendel JF, Case JS (1983) LINKAGE-1: a PASCAL computer program for the detection and analysis of genetic linkage. J Hered 74:203–204
Weeden NF, Marx G (1987) Further genetic analysis and linkage relationships of isozyme loci in the pea. J Hered 78:153–159
Weeden NF, Wolko B (1990) Linkage map for the garden pea Pisum sativum based on molecular markers. In: Genetic maps, 5th ed (O'Brien SJ, ed). Cold Spring Harbor Laboratory, Cold Spring Harbor, New York
Weeden NF, Prowidenti R, Wolko B (1991) Prx-3 is linked to sbm, the gene conferring resistance to pea seedborne mosaic virus. Pisum Genet 23:42–43
Williams JGK, Kubelik AR, Livak KJ, Rafalski J, Tingey SV (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18:6531–6535
Wolko B, Weeden NF (1990) Additional markers for chromosome 6. Pisum Newslett 22:71–74
Yu ZH, Mackill DJ, Bonman JM, Tanksley SD (1991) Tagging genes for blast resistance in rice via linkage to RFLP markers. Theor Appl Genet 81:471–476
Author information
Authors and Affiliations
Additional information
Communicated by J. S. Beckmann
Rights and permissions
About this article
Cite this article
Timmerman, G.M., Frew, T.J., Miller, A.L. et al. Linkage mapping of sbm-1, a gene conferring resistance to pea seed-borne mosaic virus, using molecular markers in Pisum sativum . Theoret. Appl. Genetics 85, 609–615 (1993). https://doi.org/10.1007/BF00220920
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00220920