Abstract
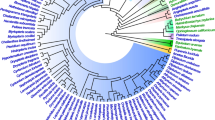
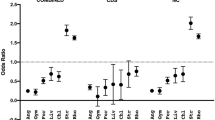
Nucleotide sequence from a region of the chloroplast genome is presented for 12 species spanning four subfamilies of the grass family. The region contains the coding sequence for the rbcL gene and the intergenic spacer between the gene coding the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase rbcL and the photosystem I gene psal. This intergenic spacer contains a pseudogene for rpl23 as well as two noncoding segments with different A+T contents. Using the sequence of rbcL a chloroplast phylogeny of this family was constructed by parsimony. Variable sites of the two noncoding segments were traced onto the phylogeny to study the dynamics of base substitution. This was also performed for the fourfold-degenerate sites of the rbcL gene. A wide variation in transversion/transition is observed between the two noncoding segments and between the noncoding DNA and the fourfold-degenerate sites of rbcL This variation is correlated with regional A+T content. As regional A+T content decreases, the ratio of transversions to transitions also decreases. Substitutions were then scored in relation to neighboring base composition. The composition of the two bases immediately flanking each substitution is highly correlated with the transversion/transition bias. When both the 5′ and 3′ flanking bases are an A or a T, transversions are observed 2.2 times as frequently as transitions. When either or both neighbors are a C or a G, the opposite trend is found; transitions are observed 1.5 times more frequently than transversions.
Similar content being viewed by others
References
Akiyama M, Maki H, Sekiguchi M, Horiuchi T (1989) A specific role of MutT protein: to prevent dG · dA mispairing in DNA replication. Proc Natl Acad Sci USA 86:3949–3952
Blake RD, Hess ST, Nicholson-Tuell J (1992) The influence of nearest neighbors on the rate and pattern of spontaneous point mutations. J Mol Evol 34:189–200
Brown WM, Prager EM, Wang A, Wilson AC (1982) Mitochondrial DNA sequences of primates: tempo and mode of evolution. J Mol Evol 18:225–239
Cabrera M, Nghiem Y, Miller JH (1988) mutM, a second mutator locus in Escherichia coli that generates G · C-T · A transversions. J Bacteriol 170:5405–5407
Clary DO, Wolstenholme DR (1987) Drosophila mitochondrial DNA: conserved sequences in the A+T-rich region and supporting evidence for a secondary structure model of the small ribosomal RNA. J Mol Evol 25:116–125
Clayton WD, Renvoize SA (1986) Genera Graminum. Grasses of the world. Her Majesty's Stationery Office, London.
Clegg MT (1993) Chloroplast gene sequences and the study of plant evolution. Proc Natl Acad Sci USA 90:363–367
DeSalle R, Freedman T, Prager EM, Wilson AC (1987) Tempo and mode of sequence evolution in mitochondrial DNA of Hawaiian Drosophila. J Mol Evol 26:157–164
Devereux J, Haeberli H, Smithies O (1984) A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res 12: 387–395
Doebley J, Durbin M, Golenberg EM, Clegg MT, Ma DP (1990) Evolutionary analysis of the large subunit of carboxylase rbcL nucleotide sequence among the grasses (Gramineae). Evolution 44: 1097–1108
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf material. Phytochem Bull 19:11–15
Duvall MR, Learn GH, Eguiarte LE, Clegg MT (1993a) Phylogenetic analysis of rbcL sequence identifies Acorus calamus as the primal extant monocotyledon. Proc Natl Acad Sci USA 90:4641–4644
Duvall MR, Chase MW, Soltis DE, Clegg MT (1993b) Phylogenetic hypotheses for the monocotyledons constructed from rbcL sequence data. Ann Miss Bot Gard 80:607–619
Gaut BS, Muse SV, Clark WD, Clegg MT (1992) Relative rates of nucleotide substitution at the rbcL locus of monocotyledonous plants. J Mol Evol 35:292–303
Gojobori T, Li WH, Grant D (1982) Patterns of nucleotide substitution in pseudogenes. J Mol Evol 18:360–369
Hiratsuka J, Shimada H, Whittier R, Ishibashi T, Sakamoto M, Mori M, Kondo C, Honji Y, Sun C-R, Meng B-Y, Li Y-Q, Kanno A, Nishizawa Y, Hirai A, Shinozaki K, Sugiura M (1989) The complete sequence of the rice (Oryza sativa) chloroplast genome: intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol Gen Genet 217:185–194
Hudson GS, Mahon JD, Anderson PA, Gibbs MJ, Badger MR, Andrews TJ, Whitfield PR (1990) Comparison of rbcL genes for the large subunit of ribulose-bisphosphate carboxylase from closely related C3 and C4 plant species. J Biol Chem 265:808–814
Jiricny J, Hughes M, Corman N, Rudkin BB (1988) A human 200-kDa protein binds selectively to DNA fragments containing G · T mismatches. Proc Natl Acad Sci USA 85:8860–8864
Jones M, Wagner R, Radman M (1987) Repair of a mismatch is influenced by the base composition of the surrounding nucleotide sequence. Genetics 115:605–610
Kramer B, Kramer W, Fritz HJ (1984) Different base/base mismatches are corrected with different efficiencies by the methyl-directed DNA mismatch-repair system of E. coli. Cell 38:879–887
Li WH, Wu CI, Luo CC (1984) Nonrandomness of point mutation as reflected in nucleotide substitutions in pseudogenes and its evolutionary implications. J Mol Evol 21:58–71
Maddison WP, Maddison DR (1992) MacClade: analysis of phylogeny and character evolution, version 3.0. Sinauer Associates, Sunderland, MA
Mendelman LV, Boosalis MS, Petruska J, Goodman MF (1989) Nearest neighbor influences on DNA polymerase insertion fidelity. J Biol Chem 264:14415–14423
Michaels ML, Cruz C, Miller JH (1990) mutA and mutC: two mutator loci in Escherichia coli that stimulate transversions. Proc Natl Acad Sci USA 87:9211–9215
Morton BR, Clegg MT (1993) A chloroplast DNA mutational hotspot and gene conversion in a noncoding region near rbcL in the grass family (Poaceae). Curr Genet 24:357–365
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Nghiem Y, Cabrera M, Cupples CG, Miller JH (1988) The mutY gene: a mutator locus in Escherichia coli that generates G · C-T · A transversions. Proc Natl Acad Sci USA 85:2709–2713
Nigro L, Solignac M, Sharp PM (1991) Mitochondrial DNA sequence divergence in the melanogaster and oriental species subgroup of Drosophila. J Mol Evol 33:156–162
Radman M, Wagner R (1986) Mismatch repair in Escherichia coli. Annu Rev Genet 20:523–538
Satta Y, Ishiwa H, Chigusa SI (1987) Analysis of nucleotide substitutions of mitochondrial DNAs in Drosophila melanogaster and its sibling species. Mol Biol Evol 4:638–650
Stephenson C, Karran P (1989) Selective binding to DNA base pair mismatches by proteins from human cells. J Biol Chem 264:21177–21182
Swofford DL (1991) PAUP: phylogenetic analysis using parsimony, version 3.0s. Computer program distributed by the Illinois Natural History Survey, Champaign
Swofford DL, Olsen GJ (1990) Phylogeny reconstruction. In: Hillis DM, Moritz C (eds) Molecular systematics. Sinauer Associates, Sunderland, MA, pp 411–501
Tajima F, Nei M (1982) Biases of the estimates of DNA divergence obtained by the restriction enzyme technique. J Mol Evol 18:115–120
Tamura K (1992) The rate and patter of nucleotide substitution in Drosophila mitochondrial DNA. Mol Biol Evol 9:814–825
Terachi T, Ogihara Y, Tsunewaki K (1987) The molecular basis of genetic diversity among cytoplasms of Triticum and Aegilops. 8. Complete nucleotide sequences of the rbcL genes encoding H- and L-type rubisco large subunits in common wheat and Ae crassa 4X. Jpn J Genet 62:375–387
Topal MD, Fresco JR (1976) Complementary base pairing and the origin of substitution mutations. Nature 263:285–289
Topal MD, DiGuiseppi SR, Sinha NK (1980) Molecular basis for substitution mutations. J Biol Chem 255:11717–11724
Wolstenholme DR, Clary DO (1985) Sequence evolution of Drosophila mitochondrial DNA. Genetics 109:725–744
Zurawski G, Clegg MT, Brown AHD (1984) The nature of nucleotide sequence divergence between barley and maize chloroplast DNA. Genetics 106:735–749
Author information
Authors and Affiliations
Additional information
Correspondence to: Brian R. Morton
Rights and permissions
About this article
Cite this article
Morton, B.R., Clegg, M.T. Neighboring base composition is strongly correlated with base substitution bias in a region of the chloroplast genome. J Mol Evol 41, 597–603 (1995). https://doi.org/10.1007/BF00175818
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00175818