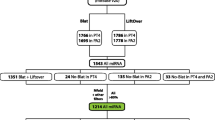
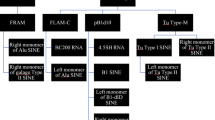
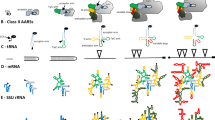
Abstract
Primate and rodent genomes are populated with hundreds of thousands copies of Alu and B1 elements dispersed by retroposition, i.e., by genomic reintegration of their reverse transcribed RNAs. These, as well as primate BC200 and rodent 4.5S RNAs, are ancestrally related to the terminal portions of 7SL RNA sequence. The secondary structure of 7SL RNA (an integral component of the signal recognition particle) is conserved from prokaryotes to distant eukaryotic species. Yet only in primates and rodents did this molecule give rise to retroposing Alu and B1 RNAs and to apparently functional BC200 and 4.5S RNAs. To understand this transition and the underlying molecular events, we examined, by comparative analysis, the evolution of RNA structure in this family of molecules derived from 7SL RNA.
RNA sequences of different simian (mostly human) and prosimian Alu subfamilies as well as rodent B1 repeats were derived from their genomic consensus sequences taken from the literature and our unpublished results (prosimian and New World Monkey). RNA secondary structures were determined by enzymatic studies (new data on 4.5S RNA are presented) and/or energy minimization analyses followed by phylogenetic comparison. Although, with the exception of 4.5S RNA, all 7SL-derived RNA species maintain the cruciform structure of their progenitor, the details of 7SL RNA folding domains are modified to a different extent in various RNA groups. Novel motifs found in retropositionally active RNAs are conserved among Alu and B1 subfamilies in different genomes. In RNAs that do not proliferate by retroposition these motifs are modified further. This indicates structural adaptation of 7SL-like RNA molecules to novel functions, presumably mediated by specific interactions with proteins; these functions were either useful for the host or served the selfish propagation of RNA templates within the host genome.
Similar content being viewed by others
Abbreviations
- FAM:
-
fossil Alu element
- FLAM:
-
free left Alu monomer
- FRAM:
-
free right Alu monomer
- L-Alu:
-
left Alu subunit
- R-Alu:
-
right Alu subunit
References
Aubert M, Scott JF, Reyner M, Monier R (1968) Rearrangement of the conformation of Escherichia coli 5S RNA. Proc Natl Acad Sci USA 61:292–299
Batzer MA, Kilroy GE, Richard PE, Shaikh TH, Desselle TD Hoppens CL, Deininger PL (1990) Structure and variability of recently inserted Alu family members. Nucleic Acids Res 18:6793–6798
Britten RJ, Baron WF, Stout BD, Davidson EH (1988) Sources and evolution of human Alu repeated sequence. Proc Natl Acad Sci USA 85:4770–4774
Brosius J (1991) Retroposons-seeds of evolution. Science 251:753
Chang D-Y, Maraia RJ (1993) A cellular protein binds B1 and Alu small cytoplasmic RNAs in vitro. J Biol Chem 268:6423–6428
Clark BFC (1978) General features and implications of primary, secondary, and tertiary structure. In: Altman S (ed) Transfer RNA. MIT Press, Cambridge, pp 1–167
D'Alessio JM (1982) RNA sequencing. IN: Rickwood D, Hames BD (eds) Gel electrophoresis of nucleic acids. IRL Press, Washington, DC, pp 177–198
Daniels GR, Deininger P (1983) A second major class of Alu family repeated DNA sequences in a primate genome. Nucleic Acids Res 11:7595–7610
Daniels GR, Fox GM, Loewensteiner D, Schmid CW, Deininger PL (1983) Species-specific homogeneity of the primate Alu family of repeated DNA sequences. Nucleic Acids Res 11:7579–7593
Deininger PL, Batzer MA, Hutchison CA, Edgell MH (1992) Master genes in mammalian repetitive DNA amplification. Trends Genet 8:307–311
Deragon J-M, Sinnett D, Labuda D (1990) Reverse transcriptase activity from human embryonal carcinoma cells NTera 2D1. EMBO J 9:3363–3369
Economou-Pachnis A, Tsichlis PN (1985) Insertion of an Alu SINE in the human homologue of the Mlvi-2 locus. Nucleic Acids Res 13:8379–8387
Goldberg YP, Rommens JM, Andrew SE, Hutchinson GB, Lin B, Theilmann J, Graham R, Glaves ML, Starr E, McDonald H, Nasir J, Schappert K, Kalchman MA, Clarke LA, Hayden MR (1993) Identification of an Alu retrotransposition event in close proximity to a strong candidate gene for Huntington's disease. Nature 362:370–373
Gundelfinger ED, Di Carlo M, Zopf D, Melli M (1984) Structure and evolution of the 7SL RNA vomponent of dignal recognition particle. EMBO J 3:2325–2332
Harada F, Kato N (1980) Nucleotide sequences of 4.58 RNAs associated with poly (A)-containing RNAs of mouse and hamster cells. Nucleic Acids Res 8:1273–1285
Hutchinson GB, Andrew SE, McDonald H, Goldberg YP, Graham R, Rommens JM, Hayden MR (1993) An Alu element retroposition in two families with Huntington disease defines a new active Alu subfamily. Nucleic Acids Res 21:3379–3383
Jagadeeswaran P, Forget BG, Weissman S (1981) Short interspersed repetitive DNA elements in eucaryotes: transposable DNA elements generated by reverse transcription of RNA Pol III transcripts? Cell 26:141–142
Jelinek WR, Toomey TP, Leinwand L, Duncan CH, Biro PA, Choudary PV, Weissman SM, Rubin CM, Houck CM, Deininger PL, Schmid CW (1980) Ubiquitous, interspersed repeated sequences in mammalian genomes. Proc Natl Acad Sci USA 77:1398–1402
Jurka J, Smith T (1988) A fundamental division in the Alu family of repeated sequences. Proc Natl Acad Sci USA 85:4775–4778
Jurka J, Zuckerkandl E (1991) Free left arms as precursor molecules in the evolution of Alu sequences. J Mol Evol 33:49–56
Jurka J, Milosavljevic A (1991) Reconstruction and analysis of human Alu genes. J Mol Evol 32:105–121
Jurka J (1993) A new subfamily of recently retroposed human Alu repeats. Nucleic Acids Res 21:2252
Krayev AS, Kramerov DA, Skryabin KG, Ryskov AP, Bayev AA, Georgiev GP (1980) The nucleotide sequence of the ubiquitous repetitive DNA sequence B1 complementary to the most abundant class of mouse fold-back RNA. Nucleic Acids Res 8:1201–1215
Labuda D, Striker G (1989) Sequence conservation in Alu evolution. Nucleic Acids Res 17:2477–2491
Labuda D, Sinnett D, Richer C, Deragon J-M, Striker G (1991) Evolution of mouse B1 repeats-7 SL RNA folding pattern conserved. J Mol Evol 32:405–414
Larsen N, Zwieb C (1991) SRP-RNA sequence alignment and secondary structure. Nucleic Acids Res 19:209–215
Makalowski W, Mitchell GA, Labuda D (1994) Trends in Genetics 10:188–193
Maraia R (1991) The subset of mouse B1 (Alu-equivalent) sequences expressed as small processed cytoplasmic transcripts. Nucleic Acids Res 19:5695–5702
Maraia RJ, Driscoll CT, Bilyeu T, Hsu K, Darlington GJ (1993) Multiple dispersed loci produce small cytoplasmic Alu RNA. Mol Cell Biol 13:4233–4241
Martignetti JA, Brosius J (1993) A neural specific RNA polymerase III product encoded by a monomeric Alu element. Proc Natl Acad Sci U S A (in press)
Matera AG, Hellman U, Hintz MF, Schmid CW (1990) Recently transposed Alu repeats result from multiple source genes. Nucleic Acids Res 18:6019–6023
Milligan JF, Groebe R, Witherell GW, Uhlenbeck OC (1987) Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res 15:8783–8798
Muratani K, Hada T, Yamamoto Y, Kaneko T, Shigeto Y, Ohue T, Furuyama J, Higashino K (1991) Inactivation of the cholinesterase gene by Alu insertion: possible mechanism for human gene transposition. Proc Natl Acad Sci 88:11315–11319
Noller HF (1984) Structure of ribosomal RNA. Annu Rev Biochem 53:119–162
Okada N (1990) Transfer RNA-like structure of the human Alu family: implications of its generation mechanism and possible functions. J Mol Evol 31:500–510
Phillips GP, Timko JL (1972) Simple method for the characterization of 5S RNA. Anal Biochem45:319–325
Poritz MA, Strub K, Walter P (1988) Human SRP RNA and E. coli 4.5S RNA contain a highly homologous structural domain. Cell 55:4–6
Quentin Y (1989) Successive waves of fixation of B1 variants in rodent lineage history. J Mol Evol 28:299–305
Quentin Y (1988) The Alu family developed through successive waves of fixation closely connected with primate lineage history. J Mol Evol 27:194–202
Quentin Y (1992a) Fusion of a free left Alu monomer and a free right Alu monomer at the origin of the Alu family in the primate genomes. Nucleic Acids Res 20:487–493
Quentin Y (1992b) Origin of the Alu family: a family of Alu-like monomers gave birth to the left and right arms of the Ain elements. Nucleic Acids Res 20:3397–3401
Reddy R (1988) Compilation of small RNA sequences. Nucleic Acids Res 16(Suppl):r71-r86
Rogers JH (1985) The origin and evolution of retroposons. Int Rev Cytol 93:187–279
Schmid CW (1993) How many source Alu? Trends Genet 9:39
Schmid CW, Shen C-KJ (1985) The evolution of interspersed repetitive DNA sequences in mammals and other vertebrates. In: MacIntyre RJ (ed) Molecular evolutionary genetics. Plenum Press, New York, pp 323–358
Schmid C, Maraia R (1992) Transcriptional regulation and transpositional selection of active SINE sequences. Curr Opin Gene Dev 2:874–882
Schoeninger LO, Jelinek WR (1986) 4–5 S RNA is encoded by hundreds of tandemly linked genes, has a short half-life, and is bonded in vivo to poly(A)-terminated RNAs in the cytoplasm of cultured mouse cells. Mol Cell Biol 6:1508–1519
Siegel V, Walter P (1988) Each of the activities of signal recognition particle (SRP) is contained within a distinct domain: analysis of biochemical mutants of SRP. Cell 52:39–49
Sinnett D, Richer C, Deragon J-M, Labuda D (1991) Alu RNA secondary structure consists of two independent 7SL RNA-like folding units. J Biol Chem 14:8675–8678
Sinnett D, Richer C, Deragon J-M, Labuda D (1992) Alu RNA transcript in human embryonal carcinoma cells. Model of post-transcriptional selection of hamster sequences. J Mol Biol 226:689–706
Tiedge H, Chen W, Brosius J (1992) Primary structure, neural-specific expression, and dendritic location of human BC200 RNA. N Neurosci 13:2382–2390
Turner DH, Sugimoto N, Jaeger JA, Longfellow CE, Freier SM, Kerzek R (1987) Improved parameters for prediction of RNA structure. Cold Spring Harbor Symp Quant Biol 52:123–133
Uhlenbeck OC, Wu HN, Sampson JR (1987) Recognition of RNA by proteins. In: Inouye M, Dudock BS (eds) molecular biology of RNA. Academic Press, Inc, pp 285–294
Ullu E, Tschudi C (1984) Alu sequences are processed 7SL RNA genes. Nature 312:171–172
Ullu E, Weiner AM (1984) Human genes and pseudogenes for the 7SL RNA component of signal recognition particle. EMBO J 3:3303–3310
VanArsdell SW, Denison RA, Bernstein LB, Weiner AM (1981) Direct repeats flank three small nuclear RNA pseudogenes in the human genome. Cell 26:11–17
Vidaud D, Vidaud M, Bahnak BR, Siguret V, Gispert Sanchez S, Laurian Y, Meter D, Goossens M, Lavergne JM (1993) Haemophilia B due to a de novo insertion of a human-specific Alu subfamily member within the coding region of the factor IX gene. Eur J Hum Genet 1:30–36
Wallace M, Andersen L, Saulino A, Gregory P, Glover T, Collins F (1991) A de novo Alu insertion results in neurofibromatosis type 1. Nature 353:864–866
Walter P, Blobel G (1982) Signal recognition particle contains a 7 S RNA essential protein translocation across the endoplasmic reticulum. Nature 299:691–698
Watson JB, Sutcliffe JG (1987) Primate brain-specific cytoplasmic transcript of the Alu repeat family. Mol Cell Biol 7:3324–3327
Weiner AM, Deininger PL, Efstratiadis A (1986) Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem 55:631–661
Willard C, Nguyen HT, Schmid CW (1987) Existence of at least three distinct Ain subfamilies. J Mol Evol 26:180–186
Zuker M (1989) Computer prediction of RNA structure. Methods Enzymol 180:262–288
Zwieb C (1985) The secondary structure of the 7SL RNA in the signal recognition particle: functional implications. Nucleic Acids Res 13:6105–6124
Zwieb C, Larsen N (1992) The signal recognition particle (SRP) database. Nucleic Acids Res 20:2207
Author information
Authors and Affiliations
Additional information
Dedicated to Dr. Robert Cedergren on the occasion of his 25th anniversary at the University of Montreal
Correspondence to: D. Labuda
Rights and permissions
About this article
Cite this article
Labuda, D., Zitkiewicz, E. Evolution of secondary structure in the family of 7SL-like RNAs. J Mol Evol 39, 506–518 (1994). https://doi.org/10.1007/BF00173420
Received:
Revised:
Issue Date:
DOI: https://doi.org/10.1007/BF00173420