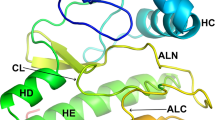
Abstract
A protein phylogenetic tree was constructed from 24 homologous proteinase inhibitor I sequences identified in the EMBUGenbank and Swiss-Prot databases and from translated amino acid data from four constitutive cDNA clones of proteinase inhibitor I characterized from potato tuber mRNA. The tree suggests that divergence of at least four paralogous proteins with functional specialization occurred at different times during the evolutionary history of the proteinase inhibitor I family. Five distinct regions in the primary structure, earlier identified by structural studies, were used to analyze the inhibitor family for hypervariability (Creighton and Darby, Trends Biochem Sci 14:319–324, 1989). Mutations did not occur with higher-than-random frequency within the proteinase binding region. When isoinhibitor, orthologous, or paralogous data subsets were subsequently analyzed the same results were obtained. Comparison of the amino acid sequences for all the known potato proteinase isoinhibitor I proteins identified ten highly variable sites. These also were distributed randomly. Thus hypervariability, which has been observed in all other serine proteinase inhibitor families to date, appears to be lacking in the proteinase inhibitor I family.
Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman BJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Bairoch A (1992) Prosite: a dictionary of sites and patterns in proteins. Nucleic Acids Res 20:(Suppl)2013–2018
Bairoch A, Boeckmann B (1992) The SWISS-PROT protein sequence data bank. Nucleic Acids Res 20:(Suppl)2019–2022
Bode W, Huber R (1991) Natural protein proteinase inhibitors and their interaction with proteinases. Eur J Biochem 204:433–451
Borriello F, Krauter KS (1991) Multiple murine α1-protease inhibitor genes show unusual evolutionary divergence. Proc Natl Acad Sci 88:9417–9421
Beuning LL, Christeller IT (1993) Isolation of a cDNA for proteinase inhibitor I. Plant Physiol 102:1061
Burks C, Cinkoskey MJ, Fischer WM, Gilna P, Hayden JED, Keen GM, Kelley M, Kristofferson D, Lawrence J (1992) GenBank. Nucleic Acids Res 20:(Suppl)2065–2069
Carrell RW, Pemberton PA, Boswell DR (1987) The serpins: evolution and adaption in a family of protease inhibitors. Cold Spring Harb Quant Biol LII:527–535
Christeller JT, Laing WA, Markwick NP, Burgess EPJ (1992) Midgut protease activities in 12 phytophagous lepidopteran larvae: dietary and protease inhibitor interactions. Insect Biochem Mol Biol 22: 735–746
Clark S (1991) Using MALIGNED to display multiple sequence alignments: emphasizing regions of conservation and divergence. Software Manual.
Cleveland TE, Thornburg RW, Ryan CA (1987) Molecular characterization of a wound-inducible inhibitor I gene from potato and the processing of its mRNA and protein. Plan Mol Biol 8:199–207
Clore GM, Gronenborn AM, Kjaer M, Poulsen FM (1987a) The determination of three-dimensional structure of barley serine proteinase inhibitor 2 by nuclear magnetic resonance, distance geometry and restrained molecular dynamics. Protein Eng 1:305–311
Clore GM, Gronenborn AM, James MNG, Kjaer M, McPhalen CA, Poulsen FM (1987b) Comparison of the solution and X-ray structures of barley proteinase inhibitor 2. Protein Eng 1:313–318
Creighton TE, Charles IG (1987) Biosynthesis, processing, and evolution of bovine pancreatic trypsin inhibitor. Cold Spring Harb Quant Biol LII:511–519
Creighton TE, Darby NJ (1989) Functional evolutionary divergence of proteolytic enzymes and their inhibitors. Trends Biochem Sci 14: 319–324
Criqui M-C, Plesse B, Dun A, Marbach J, Parmentier Y, Jamet E, Fleck J (1992) Characterization of genes expressed in mesophyll protoplasts of Nicotiana sylvestris before the re-initiation of the DNA replicational activity. Mech Dev 38:121–132
Dayhoff MO, Schwartz RM, Orcutt BC (1978) A model of evolutionary change in proteins. In: Dayhoff MO (ed) Atlas of protein sequence and structure, vol 5, supp 3. Natl Biomed Res Found, Washington, pp 345–352
Doolittle RF (1993) Convergent evolution: the need to be concise. Trends Biochem Sci 19:15–18
Frigerio F, Coda A, Pugliese L, Lionetti C, Menegatti E, Amiconi G, Schnebli HP, Ascenzi P, Bolognesi M (1992) Crystal and molecular structure of the bovine α-chymotrypsin-eglin c complex at 2.0 A resolution. J Mol Biol 225:107–123
Fujita T, Kouchi H, Ichikawa T, Syono K (1993) Isolation and characterization of a cDNA that encodes a novel proteinase inhibitor I from a tobacco genetic tumour. Plant Cell Physiol 34:137–142
Garcia-Olmedo F, Salcedo G, Sanchez-Monge M, Gomez L, Royo J, Carbonero P (1987) Plant proteinaceous inhibitors of proteinases and α-amylases. Oxford Surveys Plant Mol Cell Biol 4:275–334
Gonnet GH (1993) A tutorial introduction to computation biochemistry using Darwin. Institute for Scientific Computing, E.T.H. Zurich, Switzerland
Graham JS, Pearce G, Merryweather J, Titani K, Ericsson L, Ryan CA (1985) Wound-induced proteinase inhibitors from tomato leaves. 1. The cDNA-deduced primary structure of preinhibitor I and its post-translational processing. J Biol Chem 260:6555–6560
Green TR, Ryan CA (1972) Wound-induced proteinase inhibitor in plant leaves. Possible defense mechanism against insects. Science 175:776–777
Hickey M, King CJ (1981) 100 families of flowering plants. Cambridge University Press, Cambridge
Hilder VA, Barker RF, Samour RA, Gatehouse AMR, Gatehouse JA, Boulter D (1989) Protein and cDNA sequences of Bowman-Birk protease inhibitors from the cowpea (Vigna unguiculata Walp.) Plant Mol Biol 13:701–710
Hill RE, Hastie ND (1987) Accelerated evolution in the reactive centre regions of serine protease inhibitors. Nature 326:96–99
Hyberts SG, Wagner G (1990) Sequence-specific 1H-NMR assignments and secondary structure of eglin c. Biochemistry 29:1465–1474
Keil M, Sanchez-Serrano JJ, Willmitzer L (1989) Both wound inducible and tuber-specific expression are mediated by the promoter of a single member of the potato proteinase inhibitor II gene family. EMBO 8:1323–1330
Kjaer M, Ludvigsen S, Sorenson OW, Denys LA, Kindtler J, Poulsen FM (1987) Sequence specific assignment of the proton nuclear magnetic resonance spectrum of barley serine proteinase inhibitor 2. Carlsberg Res Commun 52:327–354
Krishnamoorthi R, YuXi Gong, Richardson M (1990) A new protein inhibitor of trypsin and activated Hageman factor from pumpkin (Cucurbita maxima) seeds. FEBS Lett 273:163–167
Laskowski M Jr, Kato I (1980) Protein inhibitors of proteinases. Ann Rev Biochem 49:593–626
Laskowski M Jr, Kato I, Ardelt W, Cook J, Denton A, Empie MW, Kohr WJ, Park SJ, Parks K, Schatzley BL, Schoenberger OL, Tashiro M, Vichot G, Wahtley HE, Wieczorek A, Wieczorek M (1987a) Ovomucoid third domain from 100 avian species: isolation, sequences, and hypervariability of enzyme-inhibitor contact residues. Biochemistry 26:202–221
Laskowski M Jr, Kato I, Kohr WJ, Park SJ, Tashiro M, Whatley HE (1987b) Positive Darwinian selection in evolution of protein inhibitors of swine proteinases. Cold Spring Harb Symp Quant Biol LII:545–553
Lee JS, Brown WE, Graham JS, Pearce G, Kox KA, Dreher TW, Ahern KG, Pearson GD, Ryan CA (1986) Molecular characterization and phylogenetic studies of a wound-inducible proteinase inhibitor I gene from Lycopersicon species. Proc Natl Acad Sci 83:7277–7281
Logemann J, Schell J, Willmitzer L (1987) Improved method for the isolation of RNA from plant tissues. Anal Biochem 163:16–20
Ludvigsen S, Shen H, Kjaer M, Madsen JC, Poulsen FM (1991) Refinement of the three-dimensional structure of barley serine proteinase inhibitor 2 and comparison with the structures in crystals. J Mol Biol 222:621–635
McPhalen CA, James MNG (1987) Crystal and molecular structure of the serine proteinase inhibitor CI-2 from barley seeds. Biochemistry 26:261–269
McPhalen CA, James MNG (1988) Structural comparison of two serine proteinase-protein inhibitor complexes: eglin c-subtilisin Carlsberg and CI-2-subtilisin Novo. Biochemistry 27:6582–6598
McPhalen CA, Svendsen I, Jonassen I, James MNG (1985) Crystal and molecular structure of the chymotrypsin inhibitor-2 from barley seeds in complex with subtilisin Novo. Proc Natl Acad Sci USA 82:7242–7246
Maier J, Muller H, Tesch R, Witt I, Holzer H (1979) Amino acid sequence of yeast proteinase B inhibitor 1 comparison with inhibitor 2. Biochem Biophys Res Commun 91:1390–1398
Margossian LJ, Federman AD, Giovannoni JJ, Fischer RL (1988) Ethylene-regulated expression of a tomato fruit ripening gene encoding a proteinase inhibitor I with a glutamic residue at the active site. Proc Natl Acad Sci USA 85:8012–8016
Melville JC, Ryan CA (1972) Chymotrypsin inhibitor I from potatoes. Large scale preparation and characterization of its subunit components. J Biol Chem 247:3445–3453
Nozawa H, Yamagata H, Aizono Y, Yoshikawa M, Iwasaki T (1989) The complete amino acid sequence of a subtilisin inhibitor from adzuki beans (Vigna angularis). J Biochem 106:1003–1008
Ogata F, Miyata T, Fujii N, Yoshida N, Noda K, Makisumi S, Ito A (1991) Purification and amino acid sequence of a Bitter Gourd inhibitor against an acid amino acid-specific endopeptidase of Streptomyces griseus. J Biol Chem 266:16715–16721
Plunkett G, Ryan CA (1980) Reduction and carboxaminomethylation of the single disulfide bond of proteinase inhibitor I from potato tubers. Effects on stability, immunological properties, and inhibitory activities. J Biol Chem 255:2752–2755
Plunkett G, Senear DF, Zuroske G, Ryan CA (1982) Proteinase inhibitors I and II from leaves of wounded tomato plants: purification and properties. Arch Biochem Biophys 213:463–472
Porter CL (1959) Taxonomy of flowering plants. WH Freeman & Co, London
Richardson M, McMillan RT, Barker RD (1976) The protomer isoinhibitors of chymotryptic inhibitor I from potatoes. Biochem Soc Trans 4:1107–1108
Richardson M, Cossins L (1974) Chymotryptic inhibitor I from potatoes: the amino acid sequences of subunits B, C, and D. FEBS Lett 45:11–13. Corrigendum: Richardson M and Cossins L (1975) FEBS Lett 52:161
Ryan CA (1978) Proteinase inhibitors in plant leaves: a biochemical model for pest-induced natural plant protection. Trends Biochem Sci 5:148–150
Ryan CA, Balls AK (1962) An inhibitor of chymotrypsin from Solanum tuberosum and its behaviour toward trypsin. Proc Natl Acad Sci USA 48:1839–1844
Schecter I, Berger A (1967) On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun 27:157–162
Schu P, Suarez Rendueles P, Wolf DH (1991) The proteinase yscB inhibitor (PBI2) gene of yeast and studies on the function of its protein product. Eur J Biochem 197:1–7
Seemuller U, Eulitz M, Fritz H, Strobel A (1980) Structure of the Elastase-Cathepsin G inhibitor of the leech Hirudo medicinalis. Hoppe-Seyler's Z Physiol Chem 361:1841–1846
Sneath PH, Sokal RR (1973) Numerical taxomnomy. Freeman, San Francisco
Svendsen IB, Boisen S, Hejgaard J (1982) Amino acid sequence of serine protease inhibitor CI-1 from barley. Homology with barley inhibitor CI-2, potato inhibitor 1, and leech eglin. Carlsberg Res Commun 47:45–53
Svendsen IB, Hejgaard J, Chavan JK (1984) Subtilisin inhibitor from seeds of broad bean (Vicia faba); purification, amino acid sequence and specificity of inhibition. Carlsberg Res Commun 49:493–502
Svendsen IB, Martin B, Jonassen I (1980) Characteristics of Hiproly barley. III. Amino acid sequences of two lysine-rich proteins. Carlsberg Res Commun 45:79–85
Swoffort DL (1990) PAUP: phylogenetic analysis using parsimony, version 3.0. Illinois Natural History Survey, Champaign, IL
Takhtajan A (1969) Flowering plants: origin and dispersal (Jeffery, C, trans). Oliver and Boyd Ltd, Edinburgh
Tashiro M, Asao T, Nakano H, Takahashi K, Kanamori M (1991) Purification and characterization of a subtilisin inhibitor from seeds of foxtail millet, Setaria italica. Agric Biol Chem 55:265–267
Wingate VPM, Broadway RM, Ryan CA (1989) Isolation and characterization of a novel, developmentally regulated proteinase inhibitor I protein and cDNA from the fruit of a wild tomato species. J Biol Chem 264:17734–17738
Wingate VPM, Ryan CA (1991) A novel fruit-expressed trypsin inhibitor I gene from a wild species of tomato. J Biol Chem 266:5188–5814
Zeng F-Y, Qian R-Q, Wang Y (1988) The amino acid sequence of a trypsin inhibitor from the seeds of Momordica charantia Linn. Cucurbitaceae. FEBS Lett 234:35–38
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Beuning, L.L., Spriggs, T.W. & Christeller, J.T. Evolution of the proteinase inhibitor I family and apparent lack of hypervariability in the proteinase contact loop. J Mol Evol 39, 644–654 (1994). https://doi.org/10.1007/BF00160410
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00160410