Summary
Metastasis and resistance to chemotherapy are common features of progressed cancers. With respect to the latter phenotype, it is thought that during tumor growth drug-resistant cells arise spontaneously at rates characteristic of the genetic alterations involved. On application of chemotherapy, such variant tumor cells are more likely to survive, and they may eventually dominate, resulting in a non-responsive malignancy. Aspects of this model have been confirmed in a number of experimental systems and in patients. In contrast to our understanding of drug resistance, steps involved in the progression to metastatic spread of tumor cells are much less well-understood.
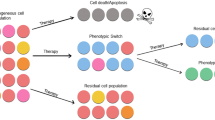
In this review we describe methodologies of quantitative genetic analysis with reference to development of drug resistance. We then describe attempts by ourselves and others to use a similar approach to investigate inetastatic properties. Based on these studies, we have proposed the quantitative ‘dynamic heterogeneity’ model of tumor metastasis, which is presented here. Using an ‘experimental’ metastasis assay and Luria-Delbruck fluctuation analysis, we determined that in murine KHT fibrosarcoma and B16 melanoma lines, metastatic’ variants with a distinct phenotype are generated at high rates. These variants are relatively anstable resulting in a dynamic equilibrium between generation and loss of metastatic variants. The metastatic ability of such a tumor population is thus dependent on the frequency of a subpopulation of metastatic variants which are turning over rapidly. This dynamic heterogeneity model is able to quantitatively provide a unifying explanation for a wide range of observations concerning tumor heterogeneity and clonal instability. Genetic mechanisms involving rapid rates have been characterized in drug-resistant variants. We speculate that similar processes may be involved in different aspects of tumor progression such as those resulting in metastasis.
Similar content being viewed by others
References
Foulds L: The experimental study of tumor progression: a review. Cancer Res 14: 327–339, 1954.
Foulds L: Neoplastic development. Academic Press, London, 1975.
Bellett AJD, Younghusband HB: Spontaneous mutagen-induced, and adenovirus-induced anchorage independent variants of mouse cells. J Cell Physiol 101: 33–48, 1979.
Thomassen DG, DeMars R: Clonal analysis of the stepwise appearance of anchorage independence and tumorigenicity in CAK, a permanent line of mouse cells. Can Res 42: 4054–4063, 1982.
Crawford BD, Barrett JC, Ts'o POP: Neoplastic conversion of preneoplastic syrian hamster cells: rate estimation by fluctuation analysis. Molec Cell Biol 3: 931–945, 1983.
Varshaver NB, Marshak MI, Shapiro NI: genetic nature of one of the traits of malignant cell transformation in vitro. Genetika (Moskra) 19: 981–987, 1983.
Shin S, Freedman VH, Risser R, Pollack R: Tumorigenicity of virus-transformed cells in nude mice is correlated specifically with anchorage independent growthin vitro Proc Natl Acad Sci USA 72: 4435–4439, 1975.
Smets LA: Cell transformation as a model for tumor induction and neoplastic growth. Biochem Biophys Acta 605: 93–111, 1980.
Ponten J: The relationship betweenin vitro transformation and tumor formationin vivo. Biochem Biophys Acta 458: 397–422, 1976.
Smith BL, Sager R: Multistep origin of tumor-forming ability in Chinese hamster embryo fibroblast cells. Can Res 42: 389–396, 1982.
Owen AH, Coffey DS, Baylin SB: Tumor cell heterogeneity: origins and implications. Bristol-Myers Cancer Symposia, Vol 4, Academic Press, New York, 1982.
Fidler IJ, Hart IR: Biological diversity in metastatic neoplasms: origins and implications. Science 217: 998–1003, 1982.
Heppner GJ, Miller BE: Tumor heterogeneity: biological implications and therapeutic consequences. Cancer Met Reviews 2: 5–23, 1983.
Nicolson GL: Generation of phenotypic diversity and progression in metastatic tumor cells. Cancer Met Reviews 3: 25–42, 1984.
Ling V, Chambers AF, Harris JF, Hill RP: Dynamic heterogeneity and metstasis. J Cell Physiol (suppl) 3: 99–103, 1984.
Nowell PC: The clonal evolution of tumor cell populations. Science 194: 23–28, 1976.
Nowell PC: Tumor progression and clonal evolution: the role of genetic instability. In: German J (ed) Chromosome mutations and neoplasia. Alan R. Liss, New York, 1983, pp 413–432.
Ponder BAJ: Genetics and cancer. Biochim Biophys Acta 605: 369–410, 1980.
Sugimura T, Sato S, Nagao M: Overlapping of carcinogens and mutagens. In: Magee PN et al (eds) Fundamentals in cancer prevention. University Park Press, Baltimore, 1976, pp 191–215.
Setlow RB: Repair deficient human disorders and cancer. Nature 271: 713–717, 1978.
Hecht F, McCaw BK: Chromosome instability syndromes. In: Mulvihill JJ, Miller RW and FraumeniJr JF (eds) Genetics of human cancer. Raven Press, New York, 1977, pp 105–123.
Fialkow PJ: Clonal origin of human tumors. Biochim Biophys Acta 458: 283–321, 1976.
Yunis JJ: The chromosomal basis of human neoplasia. Science 221: 227–236, 1983.
Vogel F: Genetics of retinoblastoma. Hum Genet 52: 1–54, 1979.
Sandberg AA: The chromosome in human cancer and leukemia. Elsevier North Holland, Inc., 1980.
Cooper GM: Cellular transforming genes. Science 218: 801–806, 1982.
Weinberg RA: Oncogenes of spontaneous and chemically induced tumors. Adv Cancer Res 36: 149–163, 1982.
Van de Woude GF, Levine AJ, Topp WC, Watson JD: Cancer cells. Vol 2 Oncogenes and viral genes. Cold Spring Harbor Laboratory, 1984.
Marx JL: What do oncogenes do? Science 223: 673–676, 1984.
Slaman DJ, deKernion JB, Verma IM, Cline MJ: Expression of cellular oncogenes in human malignancies. Science 224: 256–262, 1984.
Hunter T: Oncogenes and proto-oncogenes: how do they differ? JNCI 73: 773–786, 1984.
Thompson LH, Baker RM: Isolation of mutants of cultured mammalian cells. In: Prescott DM (ed) Methods in cell biology. Academic Press, New York, 973, Vol 6 pp 209–281.
Baker RM, Ling V: Membrane mutants of mammalian cells in culture. In: EKorn (ed) Methods in membrane biology. Plenum Publ Corp, New York, 1978, Vol 9, pp 337–384.
Ling V: Genetic basis of drug resistance in mammalian cells. In: NBruchovsky and JHGoldie (eds) Drug and hormone resistance in neoplasia. CRC Press, Miami, 1982, vol 1, pp 1–19.
Horns RC, Dower WJ, Schimke RT: Gene amplification in a leukemic patient treated with methotrexate. J Clin Oncol 2: 2–7, 1984.
Carman MD, Schornagel JH, Rivest RS, Srimatkandada A, Portlock CS, Duffy T, Bertino JR: Resistance to methotrexate due to gene amplification in a patient with acute leukemia. J Clin Oncol 2: 16–20, 1984.
Bell DR, Gerlach JH, Kartner N, Buick RN, Ling V: Detection of P-glycoprotein in ovarian cancer: a molecular marker associated with multidrug resistance. J Clin Oncol 3: 311–315, 1985.
Chu EHY, Powell SS: Selective systems in somatic cell genetics. In: HHarris and KHirschhorn (eds) Advances in human genetics. Plenum Publ Corp. New York, 1976. Vol 7, pp 189–258.
Puck TT, Kao F-T: Somatic cell genetics and its application to medicine. Ann Rev Genet 16: 225–271, 1982.
Curt GA, Clendeninn NJ, Chabner BA: Drug resistance in cancer. Cancer Treat Rep 68: 87–99, 1984.
Chan VL, Whitmore GF, Siminovitch L: Mammalian cells with altered forms of RNA polymerase II. Proc Natl Acad Sci USA 69: 3119–3124, 1972.
Ingles CJ, Guialis A, Lam J, Siminovitch L: α-amanitin resistance of RNA polymerase II in mutant Chinese hamster ovary cell lines. J Biol Chem 251: 2729–2734, 1976.
Ingles CJ, Shales M: DNA-mediated transfer of an RNA polymerase II genes: reversion of the temperature-sensitive hamster cell cycle mutants ts AF8 by mammalian DNA. Molec Cell Biol 2: 666–673, 1982.
Chan VL, Juranka P: Isolation and preliminary characterization of 9-β-D-arabinofuranosyladenine-resistant mutants of baby hamster cells. Somat Cell Genet 7: 147–160, 1981.
Andrulis IL, Siminovitch L: DNA-mediated gene transfer and β-aspartyl-hydroxamate resistance into Chinese hamster ovary cells. Proc Natl Acad Sci USA 78: 5724–5728, 1981.
Andrulis IL, Duff C, Evans-Blackler S, Worton R, Siminovitch L: Chromosomal alterations associated with overproduction of asparagine synthetase in albizziin-resistant Chinese hamster ovary cell. Molec Cell Biol 3: 391–398, 1983.
Gantt JS, Chiang C-S, Hatfield GW, Arfin S: Chinese hamster ovary cells resistant to β-aspartyl-hydroxamate contain increased levels of asparagine synthetase. J Biol Chem 255: 4808–4813, 1980.
Ling V, Thompson LH: Reduced permeability in CHO cells as a mechanism of resistance to colchicine. J Cell Physiol 83: 103–116, 1974.
Bech-Hansen NT, Till JE, Ling V: Pleiotropic phenotype of colchicine-resistant CHO cells: cross-resistance and collateral sensitivity. J Cell Physiol 88: 23–31, 1976.
Debenham PG, Kartner N, Siminovitch L, Riordan JR, Ling V: DNA-mediated transfer of multiple drug resistance and plasma membrane glycoprotein expression. Mol Cell Biol 2: 881–889, 1982.
Robertson SM, Ling V, Stanners CP: Co-amplification of double minute chromosomes, multidrug-resistance, and cell surface P-glycoprotein in DNA-mediated transformants of mouse cells. Molec Cell Biol 4: 500–506, 1984.
Kartner N, Riordan JR, Ling V: Cell surface P-glycoprotein associated with multidrug resistance in mammalian cell lines. Science 221: 1285–1288, 1983.
Riordan JR, Ling V: Genetic and biochemical characterization of multidrug resistance. In: Goldman ID (ed) The International Encyclopedia of Pharmacology and Therapeutics, Pergamon Press, (in press).
Kartner N, Shales M, Riordan JR, Ling V: Daunorubicin-resistant Chinese hamster ovary cells expressing multidrug resistance and a cell surface P-glycoprotein. Cancer Res 43: 4413–4419, 1983.
Gupta RS, Siminovitch L: The isolation and preliminary characterization of somatic cell mutants resistant to the protein synthesis inhibitor emetine. Cell 9: 213–219, 1976.
Gupta RS, Siminovitch L: The molecular basis of emetine resistance in Chinese hamster ovary cells: alteration in the 40S ribosomal subunit. Cell 10: 61–66, 1977.
Lewis WH, Wright JW: Genetic characterization of hydroxyurea-resistance in Chinese hamster ovary cells. J Cell Physiol 97: 73–85, 1978.
Lewis WH, Wright JA: Isolation of hydroxyurea-resistant CHO cells with altered levels of ribonucleotide reductase. Somat Cell Genet 5: 83–95, 1979.
Lewis WH, Srinivasan P: Chromosome-mediated gene transfer of hydroxyurea resistance and amplification of ribounucleotide reductase activity. Molec Cell Biol 3: 1053–1061, 1983.
Flintoff WF, Davidson SV, Siminovitch L: Isolation and partial characterization of three methotrexate-resistant phenotypes from CHO cells. Somat Cell Genet 2: 245–261, 1976.
Flintoff WF, Spindler SM, Siminovitch L: Genetic characterization of methotrexate-resistant CHO cells.In vitro 12: 749–757, 1976.
Milbrandt JD, Azizkhan JC, Hamlin JL: Amplification of a cloned Chinese hamster dihydrofolate reductase gene after transfer into a dihydrofolate reductase-deficient cell line. Molec Cell Biol 3: 1274–1282, 1983.
Johnston RN, Beverley SM, Schimke RT: Rapid spontaneous dihydrofolate reductase gene amplification shown by fluorescence-activated cell sorting. Proc Natl Acad Sci USA 80: 3711–3715, 1983.
Baker PM, Brunette DM, Mankovitz R, Thompson LH, Whitmore GF, Siminovitch L, Till JE: Ouabain-resistant mutants of mouse and hamster cells in culture. Cell 1: 9–21, 1974.
Kempe TD, Swyryd EA, Bruist M, Stark GR: Stable mutants of mammalian cells that overproduce the first three enzymes of pyrimidine nucleotide biosynthesis. Cell 9: 541–550, 1976.
Zieg J, Clayton CE, Ardeshir F, Giulotto E, Swyryd EA, Stark GR: Properties of single-step mutants of Syrian hamster cell lines resistant to N-(phosphonacetyl)-L-aspartate. Molec Cell Biol. 3: 2089–2098, 1983.
Cowell JK: Double minutes and homogeneously staining regions: gene amplification in mammalian cells. Ann Rev Genet 16: 21–59, 1982.
Schimke RT: Gene amplification. Cold Spring Harbor Laboratory, New York, 1982.
Luria SE, Delbruck M: Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28: 491–511, 1943.
Armitage PJ: The statistical theory of bacterial populations subject to mutations. J Roy Statist Soc B 14: 1–40, 1952.
Kondo S: A theoretical study on spontaneous mutation rate. Mutation Res 14: 365–374, 1972.
Lea DE, Coulson CA: The distribution of the numbers of mutants in bacterial populations. J Genet 49: 264–285, 1949.
Hill RP, Chambers AF, Ling V, Harris JF: Dynamic heterogeneity: Rapid generation of metastatic variants in mouse B16 melanoma cells. Science 224: 998–1001, 1984.
DeVitaJr V: The relationship between tumor mass and resistance to chemotherapy. Cancer 51: 1209–1220, 1983.
Ling V, Gerlach J, Kartner N: Multidrug resistance. Breast Cancer Res and Treat 4: 89–94, 1984.
Goldie JH, Coldman AJ: Clinical implications of the phenomenon of drug resistance. In: Bruchovsky N and Goldie JH (eds) Drug and hormone resistance in neoplasia. CRC Press, Florida, 1982, Vol II, pp 111–127.
Skipper HE: The forty-year-old mutation theory of Luria and Delbruck and its pertinence to cancer chemotherapy. Adv Canc Res 40: 331–363, 1983.
Goldie JH, Coldman AJ: A mathematical model for relating the drug sensitivity of tumors to their spontaneous mutation rate. Cancer Treat Rep 63: 1727–1733, 1979.
Chan VL, Guttman S, Juranka P: Mutator genes of baby hamster kidney cells. Molec and Cell Biol 1: 568–571, 1981.
Weinberg G, Ullman B, MartinJr DW: Mutator phenotypes in mammalian cell mutants with distinct biochemical defects and abnormal deoxyribonucleoside triphosphate pools. Proc Natl Acad Sci USA 78: 2447–2451, 1981.
Meuth M, L'Heureaux-Huard N, Trudel M: Characterization of a mutator gene in Chinese hamster ovary cells. Proc Natl Acad Sci USA 76: 6505–6509, 1979.
Drobetsky E, Meuth M: Increased mutational rates in Chinese hamster ovary cells serially selected for drug resistance. Molec Cell Biol 3: 1882–1885, 1983.
Goldberg S, Defendi V: Increased mutation rates in doubly viral transformed Chinese hamster cells. Somat Cell Genet 5: 887–895, 1979.
Gupta RS, Goldstein S: Diptheria toxin resistance in human fibroblast cell strains from normal and cancer-prone individuals. Mutation Res 73: 331–338, 1980.
Warren ST, Schultz RA, Chang C-C, Wade MH, Trosko JE: Elevated spontaneous mutation rate in Bloom syndrome fibroblasts. Proc Natl Acad Sci USA 78: 3133–3137, 1981.
Cifone MA, Fidler IJ: Increasing metastatic potential is associated with increasing genetic instability of clones isolated from murine neoplasms. Proc Natl Acad Sci USA 78: 6949–6952, 1981.
Hill RP, Bush RS: A lung-colony assay to determine the radiosensitivity of the cells of a solid tumor. Int J Radiat Biol 15: 435–444, 1969.
Fidler IJ: Selection of successive tumour lines for metastasis. Nature New Biol 242: 148–149, 1973.
Gershwin ME, Ruebner BH, Ikeda RM: Transplantation and metastasis of NB rat prostate neoplasia in congenitally athymic (nude) mice, nude mice treated with antilymphocyte sera and congenitally athymic rats. Expl Cell Biol 50: 145–154, 1982.
Kerbel RS, Man MS, Dexter D: A model of human cancer metastasis: extensive spontaneous and artificial metastasis of a human pigmented melanoma and derived variant sublines in nude mice. J Natl Can Inst 72: 93–108, 1984.
Chambers AF, Shafir R, Ling V: A model system for studying metastasis using the embryonic chick. Cancer Res 42: 4018–4025, 1982.
Kripke ML, Gruys E, Fidler IJ: Metastatic heterogeneity of cells from an utraviolet light-induced murine fibrosarcoma of recent origin. Cancer Res 38: 2962–2967, 1978.
Fidler IJ, Cifone MA: Properties of metastatic and non-metastatic cloned subpopulations of an ultraviolet-light-induced murine fibrosarcoma of recent origin. Amer J Path 97: 633–648, 1979.
Welch DR, Neri A, Nicolson GL: Comparison of ‘spontaneous’ and ‘experimental’ metastasis using rat 13762 mammary adenocarcinoma metastatic cell clones. Inv Metast 3: 65–80, 1983.
Stackpole CW: Distinct lung-colonizing and lung-metastasizing cell populations in B16 mouse melanoma. Nature 289: 798–800, 1981.
Sweeney FL, Pot-Deprun J, Poupon M-F, Chourouliukov I: Heterogeneity of the growth and metastatic behavior of cloned cell lines from a primary rhabdomyosarcoma. Cancer Res 42: 3776–3782, 1982.
Price JE, Carr D, Jones LD, Messer P, Tarim D: Experimental analysis of factors affecting metastatic spread using naturally occurring tumours. Inv Metast 2: 77–112, 1982.
Glaves D: Correlation between circulating cancer cells and incidence of metastases. Br J Cancer 48: 665–673, 1983.
Suzuki N: Spontaneous versus artificial lung metastasis: Discrepant effect of whole-body irradiation in NFSA2ALM and NFSA1SLM tumor systems. J Natl Cancer Inst 71: 835–839, 1983.
Ramshaw IR, Carlsen S, Wang HC, Badenoch-Jones P: The use of cell fusion to analyse factors involved in tumour cell metastasis. Int J Cancer 32: 471–478, 1983.
Sidebottom E, Clark SR: Cell fusion segregates progressive growth from metastasis. Br J Cancer 47: 399–406, 1983.
Hart IR: Tumor cell hybridization and neoplastic progression. In: GLNicolson and LMilas (eds) Cancer invasion and metastases: biologic and therapeutic aspects. Raven Press, New York, 1984, pp 133–143.
DeBaetselier P, Gorelik E, Eshhar Z, Ron Y, Katzav S, Feldman M, Segal S: Metastatic properties conferred on nonmetastatic tumors by hybridization of spleen B-lymphocytes with plasmacytoma cells. J Natl Cancer Inst 67: 1079–1087, 1981.
DeBaetselier P, Roos E, Brys L, Ranels L, Gobert M, Dekegel D, Segal S, Feldman M: Non-metastatic tumor cells acquire metastatic properties following somatic hybridization with normal cells. Cancer Metast Rev 3: 5–24, 1984.
Kerbel RS, Dennis JW, Lagarde AE, Frost P: Tumor progression in metastasis: an experimental approach using lectin resistant tumor variants. Cancer Metast Rev 1: 99–40, 1982.
Kerbel RS, Lagarde AE, Dennis JW, Donaghue TP: Spontaneous fusion in vivo between normal host and tumour cells: possible contribution to tumor progression and metastasis studied with a lectin-resistant mutant tumor. Molec Cell Biol 3: 523–538, 1983.
Larizza L, Schirrmacher V: Somatic cell fusion as a source of genetic rearrangement leading to metastatic variants. Cancer Metast Rev 3: 193–222, 1984.
Chambers AF, Ling V: Selection for experimental metastatic ability of heterologous tumor cells in the chick embryo after DNA-mediated transfer. Cancer Res 44: 3970–3975, 1984.
Chambers AF, Wilson S: Cells transformed with ats viralsrc mutant are temperature sensitive forin vitro growth. Mol Cell Biol 5: 728–733, 1985.
Eccles SA, Marshall C, Vousden K, Purvies H: Enhanced metastatic capacity of mouse mammary carcinoma cells transfected with H-ras. In: Hellman K and Eccles SA (eds) Treatment of metastasis. Problems and Prospects. Taylor and Francis, London, 1985.
Thorgeirsson UP, Turpeenniemi-Hujanen T, Williams JE, Westin EH, Heilman CA, Talmadge JE, Liotta LA: NIH/3T3 cells transfected with human tumor DNA containing activated ras oncogenes express the metastatic phenotype in nude mice. Molec Cell Biol 5: 259–262, 1985.
Harris JF, Chambers AF, Hill RP, Ling V: Metastatic variants are generated spontaneously at a high rate in mouse KHT tumor. Proc Natl Acad Sci 79: 5547–5551, 1982.
Chambers AF, Harris JF, Ling V, Hill RP: Rapid phenotype variation in cells derived from lung metastases of KHT fibrosarcoma. Inv Metast 4: 225–237, 1984.
Fidler IJ, Kripke ML: Metastasis results from pre-existing variant cells within a malignant tumor. Science 197: 893–895, 1977.
Dexter DL, Kowalski HM, Blazar BA, Fligiel Z, Vogel R, Heppner GH: Heterogeneity of tumor cells from a single mouse mammary tumor. Cancer Res 38: 3174–3181, 1978.
Nicolson GL, Brunson KW, Fidler IJ: Specificity of arrest, survival and growth of selected metastatic variant cell lines. Cancer Res 38: 4105–4111, 1978.
Suzuki N, Withers HR, Koehler MW: Heterogeneity and variability of artificial lung colony-forming ability among clones from mouse fibrosarcoma. Cancer Res 38: 3349–3351, 1978.
Cifone MA, Kripke ML, Fidler IJ: Growth rate and chromosome number of tumor cell lines with different metastatic potential. J Supramol Struct 11: 467–476, 1979.
Fidler IJ, Cifone MA: Properties of metastatic and non-metastatic cloned subpopulations of an ultra-violet-light-induced murine fibrosarcoma of recent origin. Am J Pathol 97: 633–648, 1979.
Schirrmacher V, Shantz G, Claver K, Komitowski D, Zimmerman HP, Lohmann-Mathes ML: Tumor metastases and cell-mediated immunity in a model system in DBA/2 mice. I Tumor invasivenessin vitro and metastasis formationin vivo. Int J Can 23: 233–244, 1979.
Chambers AF, Hill RP, Ling V: Tumor heterogeneity and stability of the metastatic phenotype of mouse KHT sarcoma cells. Cancer Res 41: 1368–1372, 1981.
Fidler IJ, Hart IR: The origin of metastatic heterogeneity in tumors. Eur J Cancer 17: 487–494, 1981.
Poste G, Doll J, Fidler IJ: Interactions between clonal subpopulations affect the stability of the metastatic phenotype in polyclonal populations of B16 melanoma cells. Proc Natl Acad Sci 78: 6226–6230, 1981.
Neri A, Welch D, Kawaguchi T, Nicolson GL: Development and biologic properties of malignant cell sublines and clones of a spontaneously metastasizing rat mammary adenocarcinoma. JNCI 68: 507–517, 1982.
Talmadge JE: The selective nature of metastasis. Cancer Metast Rev 2: 25–40, 1983.
Weiss L: Dynamic aspects of cancer cell populations in metastasis. Am J Path 97: 601–608, 1979.
Weiss L: Random and nonrandom processes in metastasis and metastatic inefficiency. Inv Metast 3: 193–207, 1983.
Poste G, Greig R: On the genesis and regulation of cellular heterogeneity in malignant tumours. Inv Metast 2: 137–176, 1982.
Heppner G, Miller BE, Miller FR: Tumor subpopulation interactions in neoplasms. Biochim Biophys Acta 695: 215–226, 1984.
Turner GA: Surface properties of the metastatic cell. Invasion Metastasis 2: 197–216, 1982.
Nicolson GL: Cancer Metastasis: organ colonization and the cell-surface properties of malignant cells. Biochim Biophy Acta 695: 113–176, 1982.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Ling, V., Chambers, A.F., Harris, J.F. et al. Quantitative genetic analysis of tumor progression. Cancer Metast Rev 4, 173–192 (1985). https://doi.org/10.1007/BF00050694
Issue Date:
DOI: https://doi.org/10.1007/BF00050694